Difference between revisions of "CatE"
(→Expression and regulation) |
|||
| Line 89: | Line 89: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | + | * '''Operon:''' ''[[catD]]-[[catE]]'' {{PubMed|16872404}} | |
| − | * '''Operon:''' ''[[catD]]-[[catE]]'' | ||
* '''[[Sigma factor]]:''' | * '''[[Sigma factor]]:''' | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| + | ** strong induction by catechol {{PubMed|16872404}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| Line 120: | Line 120: | ||
=References= | =References= | ||
| − | <pubmed>17407181,, </pubmed> | + | <pubmed>17407181,16872404 , </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 19:52, 22 May 2010
- Description: catechol 2,3-dioxygenase
| Gene name | catE |
| Synonyms | yfiE |
| Essential | no |
| Product | catechol 2,3-dioxygenase |
| Function | essential for the viability in the presence of catechol |
| MW, pI | 31 kDa, 5.302 |
| Gene length, protein length | 855 bp, 285 aa |
| Immediate neighbours | catD, yfiF |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 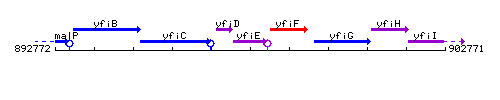
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU08240
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: P54721
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- strong induction by catechol PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Van Duy Nguyen, Carmen Wolf, Ulrike Mäder, Michael Lalk, Peter Langer, Ulrike Lindequist, Michael Hecker, Haike Antelmann
Transcriptome and proteome analyses in response to 2-methylhydroquinone and 6-brom-2-vinyl-chroman-4-on reveal different degradation systems involved in the catabolism of aromatic compounds in Bacillus subtilis.
Proteomics: 2007, 7(9);1391-408
[PubMed:17407181]
[WorldCat.org]
[DOI]
(P p)
Le Thi Tam, Christine Eymann, Dirk Albrecht, Rabea Sietmann, Frieder Schauer, Michael Hecker, Haike Antelmann
Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis.
Environ Microbiol: 2006, 8(8);1408-27
[PubMed:16872404]
[WorldCat.org]
[DOI]
(P p)