Difference between revisions of "HutG"
| Line 89: | Line 89: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | |||
* '''Operon:''' ''[[hutP]]-[[hutH]]-[[hutU]]-[[hutI]]-[[hutG]]-[[hutM]]'' [http://www.ncbi.nlm.nih.gov/sites/entrez/10746760 PubMed] | * '''Operon:''' ''[[hutP]]-[[hutH]]-[[hutU]]-[[hutI]]-[[hutG]]-[[hutM]]'' [http://www.ncbi.nlm.nih.gov/sites/entrez/10746760 PubMed] | ||
| − | * '''[[Sigma factor]]:''' [[SigA]] | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|8071225}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| + | ** repressed by glucose ([[CcpA]]) {{PubMed|8071225}} | ||
| + | ** induced by histidine ([[HutP]]) {{PubMed|8071225}} | ||
| + | ** repressed during growth in the presence of branched chain amino acids ([[CodY]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/8682780 PubMed] | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| + | ** [[CcpA]]: transcription repression {{PubMed|8071225}} | ||
| + | ** [[CodY]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/8682780 PubMed] | ||
| + | ** [[HutP]]: transcriptional antitermination at a protein-dependent [[RNA switch]] located between ''[[hutP]]'' and ''[[hutH]]'' {{PubMed|8071225}} | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| Line 120: | Line 125: | ||
=References= | =References= | ||
| − | <pubmed> | + | <pubmed>10746760 ,8682780 , 8071225</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 19:08, 8 February 2010
- Description: formiminoglutamate hydrolase
| Gene name | hutG |
| Synonyms | |
| Essential | no |
| Product | formiminoglutamate hydrolase |
| Function | histidine utilization |
| Metabolic function and regulation of this protein in SubtiPathways: His | |
| MW, pI | 34 kDa, 5.439 |
| Gene length, protein length | 957 bp, 319 aa |
| Immediate neighbours | hutI, hutM |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 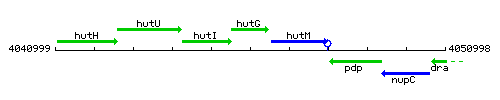
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU39380
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: N-formimidoyl-L-glutamate + H2O = L-glutamate + formamide (according to Swiss-Prot)
- Protein family: arginase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: P42068
- KEGG entry: [3]
- E.C. number: 3.5.3.8
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Ken-Ichi Yoshida, Izumi Ishio, Eishi Nagakawa, Yoshiyuki Yamamoto, Mami Yamamoto, Yasutaro Fujita
Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome.
Microbiology (Reading): 2000, 146 ( Pt 3);573-579
[PubMed:10746760]
[WorldCat.org]
[DOI]
(P p)
S H Fisher, K Rohrer, A E Ferson
Role of CodY in regulation of the Bacillus subtilis hut operon.
J Bacteriol: 1996, 178(13);3779-84
[PubMed:8682780]
[WorldCat.org]
[DOI]
(P p)
L V Wray, S H Fisher
Analysis of Bacillus subtilis hut operon expression indicates that histidine-dependent induction is mediated primarily by transcriptional antitermination and that amino acid repression is mediated by two mechanisms: regulation of transcription initiation and inhibition of histidine transport.
J Bacteriol: 1994, 176(17);5466-73
[PubMed:8071225]
[WorldCat.org]
[DOI]
(P p)