Difference between revisions of "RbfA"
| Line 88: | Line 88: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[ylxS]]-[[nusA]]-[[ylxR]]-[[ylxQ]]-[[infB]]-[[ylxP]]-[[rbfA]]'' {{PubMed|8491709}} |
| − | * '''[[Sigma factor]]:''' | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|8491709}} |
| + | |||
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 97: | Line 98: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
| Line 119: | Line 120: | ||
=References= | =References= | ||
| + | <pubmed>8491709 11948165</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 19:20, 16 October 2009
- Description: ribosome-binding factor A
| Gene name | rbfA |
| Synonyms | ymxE, ylxO |
| Essential | no |
| Product | ribosome-binding factor A |
| Function | translation |
| MW, pI | 13 kDa, 9.338 |
| Gene length, protein length | 351 bp, 117 aa |
| Immediate neighbours | ylxP, truB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 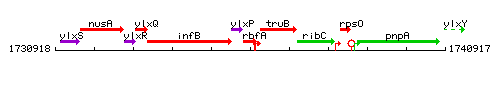
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU16650
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: rbfA family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P32731
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References