Difference between revisions of "MifM"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' ribosome-nascent chain sensor of membrane protein biogenesis <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''mifM'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' yqzJ'' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || sensor of [[SpoIIIJ]] activity |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || control of ''[[yqjG]]'' translation |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 11 kDa, 5.829 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 11 kDa, 5.829 | ||
| Line 52: | Line 52: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' acts as a kind of "leader peptide" that does or does not allow translation through a hairpin structure in the ''[[yqjG]]'' mRNA {{PubMed|19779460}} |
* '''Protein family:''' | * '''Protein family:''' | ||
| Line 70: | Line 70: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''Interactions:''' the C-terminus of MifM interacts with the ribosomal polypeptide exit tunnel {{PubMed|19779460}} |
| − | * '''Localization:''' | + | * '''Localization:''' membrane {{PubMed|19779460}} |
=== Database entries === | === Database entries === | ||
| Line 117: | Line 117: | ||
=References= | =References= | ||
| − | + | <pubmed> 19779460 </pubmed> | |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 18:20, 26 September 2009
- Description: ribosome-nascent chain sensor of membrane protein biogenesis
| Gene name | mifM |
| Synonyms | yqzJ |
| Essential | no |
| Product | sensor of SpoIIIJ activity |
| Function | control of yqjG translation |
| MW, pI | 11 kDa, 5.829 |
| Gene length, protein length | 285 bp, 95 aa |
| Immediate neighbours | polY1, yqjG |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 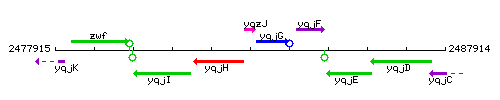
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU23880
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: acts as a kind of "leader peptide" that does or does not allow translation through a hairpin structure in the yqjG mRNA PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions: the C-terminus of MifM interacts with the ribosomal polypeptide exit tunnel PubMed
- Localization: membrane PubMed
Database entries
- Structure:
- UniProt: Q7WY64
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References