Difference between revisions of "Noc"
(→Expression and regulation) |
|||
| Line 88: | Line 88: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[thdF]]-[[gidA]]-[[rsmG]]-[[noc]]'' |
* '''[[Sigma factor]]:''' | * '''[[Sigma factor]]:''' | ||
| Line 96: | Line 96: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
Revision as of 10:44, 13 July 2009
- Description: DNA-binding protein, spatial regulator of cell division to protect the nucleoid, and timing device with an important role in the coordination of chromosome segregation and cell division
| Gene name | noc |
| Synonyms | yyaA |
| Essential | no |
| Product | effector of nucleoid occlusion |
| Function | control of cell division |
| MW, pI | 32 kDa, 5.812 |
| Gene length, protein length | 849 bp, 283 aa |
| Immediate neighbours | yyaB, gidB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 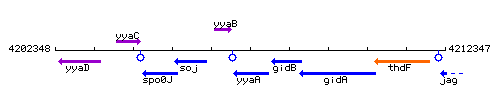
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU40990
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: binds specific sites on the chromosome PubMed
- Protein family: ParB family (according to Swiss-Prot)
- Paralogous protein(s): Spo0J
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- Swiss prot entry: P37524
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion: available in the Jeff Errington lab
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Jeff Errington, Newcastle, UK Homepage
Your additional remarks
References
Ling Juan Wu, Shu Ishikawa, Yoshikazu Kawai, Taku Oshima, Naotake Ogasawara, Jeff Errington
Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation.
EMBO J: 2009, 28(13);1940-52
[PubMed:19494834]
[WorldCat.org]
[DOI]
(I p)
Yoshikazu Kawai, Naotake Ogasawara
Bacillus subtilis EzrA and FtsL synergistically regulate FtsZ ring dynamics during cell division.
Microbiology (Reading): 2006, 152(Pt 4);1129-1141
[PubMed:16549676]
[WorldCat.org]
[DOI]
(P p)
Ling Juan Wu, Jeff Errington
Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis.
Cell: 2004, 117(7);915-25
[PubMed:15210112]
[WorldCat.org]
[DOI]
(P p)
Jörg Sievers, Brian Raether, Marta Perego, Jeff Errington
Characterization of the parB-like yyaA gene of Bacillus subtilis.
J Bacteriol: 2002, 184(4);1102-11
[PubMed:11807071]
[WorldCat.org]
[DOI]
(P p)