Difference between revisions of "Ctc"
(→Expression and regulation) |
|||
| Line 91: | Line 91: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | |||
| − | * ''' | + | * '''Operon:''' |
| + | ** ''[[gcaD]]-[[prs]]-[[ctc]]'' [http://www.ncbi.nlm.nih.gov/pubmed/8522540 PubMed] | ||
| + | ** ''[[ctc]]'' | ||
| − | * '''Regulation:''' induced by stress ([[SigB]]) [http://www.ncbi.nlm.nih.gov/pubmed/15805528 PubMed] | + | * '''[[Sigma factor]]:''' |
| + | ** ''[[gcaD]]'': [[SigA]] [http://www.ncbi.nlm.nih.gov/pubmed/8522540 PubMed] | ||
| + | ** ''[[ctc]]'': [[SigB]] | ||
| + | |||
| + | * '''Regulation:''' induced by stress ([[SigB]]) [http://www.ncbi.nlm.nih.gov/pubmed/15805528 PubMed] | ||
* '''Regulatory mechanism:''' alternative [[sigma factor]], [[SigB]] | * '''Regulatory mechanism:''' alternative [[sigma factor]], [[SigB]] | ||
Revision as of 19:22, 4 July 2009
- Description: general stress protein, similar to L25
| Gene name | ctc |
| Synonyms | |
| Essential | no |
| Product | ribosomal protein |
| Function | unknown |
| Metabolic function and regulation of this protein in SubtiPathways: Nucleotides (regulation) | |
| MW, pI | 21 kDa, 4.216 |
| Gene length, protein length | 612 bp, 204 aa |
| Immediate neighbours | prs, spoVC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 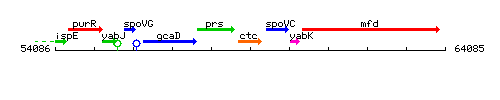
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU00520
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: Ctc subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions: binds 5S rRNA PubMed
- Localization: in the ribosome (large subunit) PubMed
Database entries
- Structure:
- Swiss prot entry: P14194
- KEGG entry: [3]
- E.C. number:
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulatory mechanism: alternative sigma factor, SigB
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody: available in Stülke lab
Labs working on this gene/protein
Your additional remarks
References
G M Gongadze, A P Korepanov, A V Korobeinikova, M B Garber
Bacterial 5S rRNA-binding proteins of the CTC family.
Biochemistry (Mosc): 2008, 73(13);1405-17
[PubMed:19216708]
[WorldCat.org]
[DOI]
(I p)
Dirk Höper, Uwe Völker, Michael Hecker
Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis.
J Bacteriol: 2005, 187(8);2810-26
[PubMed:15805528]
[WorldCat.org]
[DOI]
(P p)
Matthias Schmalisch, Ines Langbein, Jörg Stülke
The general stress protein Ctc of Bacillus subtilis is a ribosomal protein.
J Mol Microbiol Biotechnol: 2002, 4(5);495-501
[PubMed:12432960]
[WorldCat.org]
(P p)
I Hilden, B N Krath, B Hove-Jensen
Tricistronic operon expression of the genes gcaD (tms), which encodes N-acetylglucosamine 1-phosphate uridyltransferase, prs, which encodes phosphoribosyl diphosphate synthetase, and ctc in vegetative cells of Bacillus subtilis.
J Bacteriol: 1995, 177(24);7280-4
[PubMed:8522540]
[WorldCat.org]
[DOI]
(P p)
C L Truitt, E A Weaver, W G Haldenwang
Effects on growth and sporulation of inactivation of a Bacillus subtilis gene (ctc) transcribed in vitro by minor vegetative cell RNA polymerases (E-sigma 37, E-sigma 32).
Mol Gen Genet: 1988, 212(1);166-71
[PubMed:2836704]
[WorldCat.org]
[DOI]
(P p)
C Ray, M Igo, W Shafer, R Losick, C P Moran
Suppression of ctc promoter mutations in Bacillus subtilis.
J Bacteriol: 1988, 170(2);900-7
[PubMed:3123466]
[WorldCat.org]
[DOI]
(P p)
M M Igo, R Losick
Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis.
J Mol Biol: 1986, 191(4);615-24
[PubMed:3100810]
[WorldCat.org]
[DOI]
(P p)
J F Ollington, W G Haldenwang, T V Huynh, R Losick
Developmentally regulated transcription in a cloned segment of the Bacillus subtilis chromosome.
J Bacteriol: 1981, 147(2);432-42
[PubMed:6790515]
[WorldCat.org]
[DOI]
(P p)
W G Haldenwang, R Losick
A modified RNA polymerase transcribes a cloned gene under sporulation control in Bacillus subtilis.
Nature: 1979, 282(5736);256-60
[PubMed:116131]
[WorldCat.org]
[DOI]
(P p)