Difference between revisions of "MraY"
| Line 84: | Line 84: | ||
* '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/Q03521 Q03521] | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/Q03521 Q03521] | ||
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU15190] |
* '''E.C. number:''' [http://www.expasy.org/enzyme/2.7.8.13 2.7.8.13] | * '''E.C. number:''' [http://www.expasy.org/enzyme/2.7.8.13 2.7.8.13] | ||
Revision as of 03:14, 25 June 2009
- Description: phospho-N-acetylmuramoyl-pentapeptide-transferase (meso-2,6-diaminopimelate)
| Gene name | mraY |
| Synonyms | |
| Essential | yes PubMed |
| Product | phospho-N-acetylmuramoyl-pentapeptide-transferase (meso-2,6-diaminopimelate) |
| Function | peptidoglycan precursor biosynthesis |
| Metabolic function and regulation of this protein in SubtiPathways: Cell wall | |
| MW, pI | 35 kDa, 8.966 |
| Gene length, protein length | 972 bp, 324 aa |
| Immediate neighbours | murE, murD |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 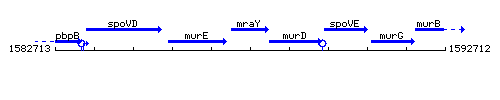
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU15190
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: UDP-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala) + undecaprenyl phosphate = UMP + Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-diphosphoundecaprenol (according to Swiss-Prot)
- Protein family: MraY subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- Swiss prot entry: Q03521
- KEGG entry: [3]
- E.C. number: 2.7.8.13
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Yi Zheng, Douglas K Struck, Thomas G Bernhardt, Ry Young
Genetic analysis of MraY inhibition by the phiX174 protein E.
Genetics: 2008, 180(3);1459-66
[PubMed:18791230]
[WorldCat.org]
[DOI]
(P p)
Bayan Al-Dabbagh, Xavier Henry, Meriem El Ghachi, Geneviève Auger, Didier Blanot, Claudine Parquet, Dominique Mengin-Lecreulx, Ahmed Bouhss
Active site mapping of MraY, a member of the polyprenyl-phosphate N-acetylhexosamine 1-phosphate transferase superfamily, catalyzing the first membrane step of peptidoglycan biosynthesis.
Biochemistry: 2008, 47(34);8919-28
[PubMed:18672909]
[WorldCat.org]
[DOI]
(I p)
Ahmed Bouhss, Amy E Trunkfield, Timothy D H Bugg, Dominique Mengin-Lecreulx
The biosynthesis of peptidoglycan lipid-linked intermediates.
FEMS Microbiol Rev: 2008, 32(2);208-33
[PubMed:18081839]
[WorldCat.org]
[DOI]
(P p)
Ahmed Bouhss, Muriel Crouvoisier, Didier Blanot, Dominique Mengin-Lecreulx
Purification and characterization of the bacterial MraY translocase catalyzing the first membrane step of peptidoglycan biosynthesis.
J Biol Chem: 2004, 279(29);29974-80
[PubMed:15131133]
[WorldCat.org]
[DOI]
(P p)
M A Lehrman
A family of UDP-GlcNAc/MurNAc: polyisoprenol-P GlcNAc/MurNAc-1-P transferases.
Glycobiology: 1994, 4(6);768-71
[PubMed:7734839]
[WorldCat.org]
[DOI]
(P p)
R A Daniel, J Errington
DNA sequence of the murE-murD region of Bacillus subtilis 168.
J Gen Microbiol: 1993, 139(2);361-70
[PubMed:8436954]
[WorldCat.org]
[DOI]
(P p)