Difference between revisions of "YumB"
| Line 42: | Line 42: | ||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| − | {{SubtiWiki category|[[proteins of unknown function]]}} | + | {{SubtiWiki category|[[proteins of unknown function]]}}, |
| + | {{SubtiWiki category|[[sporulation proteins]]}} | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
Revision as of 10:15, 21 November 2012
- Description: unknown
| Gene name | yumB |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | unknown |
| Gene expression levels in SubtiExpress: yumB | |
| MW, pI | 44 kDa, 6.616 |
| Gene length, protein length | 1218 bp, 406 aa |
| Immediate neighbours | yuiA, yumC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 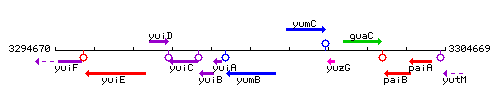
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
proteins of unknown function, sporulation proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU32100
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: O05267
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
The gene is annotated in KEGG as an ortholog of NADH dehydrogenase EC 1.6.99.3. No EC annotation is available in Swiss-Prot. In MetaCyc it is marked as “similar to NADH dehydrogenase”. In the paper by Gyan et al. PubMed, the authors tested the growth of three B. subtilis genes potentially responsible for EC 1.6.99.3 (yumB, yjlD, and yutJ). Only yjlD- mutant showed a growth defect on the LB media, while the other two grew as well as wild type. PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Tzu-Lin Hsiao, Olga Revelles, Lifeng Chen, Uwe Sauer, Dennis Vitkup
Automatic policing of biochemical annotations using genomic correlations.
Nat Chem Biol: 2010, 6(1);34-40
[PubMed:19935659]
[WorldCat.org]
[DOI]
(I p)
Smita Gyan, Yoshihiko Shiohira, Ichiro Sato, Michio Takeuchi, Tsutomu Sato
Regulatory loop between redox sensing of the NADH/NAD(+) ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis.
J Bacteriol: 2006, 188(20);7062-71
[PubMed:17015645]
[WorldCat.org]
[DOI]
(P p)