Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' penicillin-binding protein 5, D-alanyl-D-alanine carboxypeptidase <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''dacB'' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || penicillin-binding protein 5, D-alanyl-D-alanine carboxypeptidase |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || carboxypeptidase |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 42 kDa, 9.725 |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 1146 bp, 382 aa |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[spmA]]'', ''[[ypuI]]'' |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS: | + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB14251]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' |
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |colspan="2" | '''Genetic context''' <br/> [[Image:dacB_context.gif]] |
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| Line 35: | Line 35: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU23190 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| Line 41: | Line 41: | ||
=== Database entries === | === Database entries === | ||
| − | * '''DBTBS entry:''' | + | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/dacB-spmAB.html] |
| − | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+ | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10527] |
=== Additional information=== | === Additional information=== | ||
| Line 52: | Line 52: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' Preferential cleavage: (Ac)(2)-L-Lys-D-Ala-|-D-Ala (according to Swiss-Prot) |
| − | * '''Protein family:''' | + | * '''Protein family:''' peptidase S11 family (according to Swiss-Prot) |
| − | * '''Paralogous protein(s):''' | + | * '''Paralogous protein(s):''' |
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 78: | Line 78: | ||
* '''Structure:''' | * '''Structure:''' | ||
| − | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/ | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P35150 P35150] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu: | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU23190] |
* '''E.C. number:''' | * '''E.C. number:''' | ||
| Line 88: | Line 88: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[dacB]]-[[spmA]]-[[spmB]]'' |
| − | * '''[[Sigma factor]]:''' | + | * '''[[Sigma factor]]:''' [[SigE]] [http://www.ncbi.nlm.nih.gov/sites/entrez/15758244 Link to a summary] |
| − | * '''Regulation:''' | + | * '''Regulation:''' expressed during sporulation in the mother cell [http://www.ncbi.nlm.nih.gov/sites/entrez/15758244 Link to a summary] |
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
| Line 118: | Line 118: | ||
=References= | =References= | ||
| − | <pubmed> | + | <pubmed>10498740,9864321,1548223,7642500,14731276,19542328 , </pubmed> |
Revision as of 15:48, 2 July 2009
- Description: penicillin-binding protein 5, D-alanyl-D-alanine carboxypeptidase
| Gene name | dacB |
| Synonyms | |
| Essential | no |
| Product | penicillin-binding protein 5, D-alanyl-D-alanine carboxypeptidase |
| Function | carboxypeptidase |
| MW, pI | 42 kDa, 9.725 |
| Gene length, protein length | 1146 bp, 382 aa |
| Immediate neighbours | spmA, ypuI |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 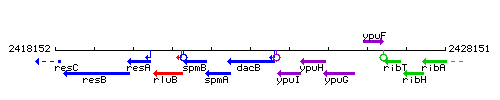
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU23190
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Preferential cleavage: (Ac)(2)-L-Lys-D-Ala-|-D-Ala (according to Swiss-Prot)
- Protein family: peptidase S11 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- Swiss prot entry: P35150
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation: expressed during sporulation in the mother cell Link to a summary
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Ralf Moeller, Peter Setlow, Günther Reitz, Wayne L Nicholson
Roles of small, acid-soluble spore proteins and core water content in survival of Bacillus subtilis spores exposed to environmental solar UV radiation.
Appl Environ Microbiol: 2009, 75(16);5202-8
[PubMed:19542328]
[WorldCat.org]
[DOI]
(I p)
Dirk-Jan Scheffers, Laura J F Jones, Jeffery Errington
Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis.
Mol Microbiol: 2004, 51(3);749-64
[PubMed:14731276]
[WorldCat.org]
[DOI]
(P p)
D L Popham, J Meador-Parton, C E Costello, P Setlow
Spore peptidoglycan structure in a cwlD dacB double mutant of Bacillus subtilis.
J Bacteriol: 1999, 181(19);6205-9
[PubMed:10498740]
[WorldCat.org]
[DOI]
(P p)
D L Popham, M E Gilmore, P Setlow
Roles of low-molecular-weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties.
J Bacteriol: 1999, 181(1);126-32
[PubMed:9864321]
[WorldCat.org]
[DOI]
(P p)
D L Popham, B Illades-Aguiar, P Setlow
The Bacillus subtilis dacB gene, encoding penicillin-binding protein 5*, is part of a three-gene operon required for proper spore cortex synthesis and spore core dehydration.
J Bacteriol: 1995, 177(16);4721-9
[PubMed:7642500]
[WorldCat.org]
[DOI]
(P p)
C E Buchanan, M L Ling
Isolation and sequence analysis of dacB, which encodes a sporulation-specific penicillin-binding protein in Bacillus subtilis.
J Bacteriol: 1992, 174(6);1717-25
[PubMed:1548223]
[WorldCat.org]
[DOI]
(P p)