PurA
- Description: adenylosuccinate synthetase
| Gene name | purA |
| Synonyms | |
| Essential | no |
| Product | adenylosuccinate synthetase |
| Function | purine biosynthesis |
| Metabolic function and regulation of this protein in SubtiPathways: Purine salvage, Purine synthesis, Nucleotides (regulation) | |
| MW, pI | 47 kDa, 5.459 |
| Gene length, protein length | 1290 bp, 430 aa |
| Immediate neighbours | trnY-Lys, yycE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 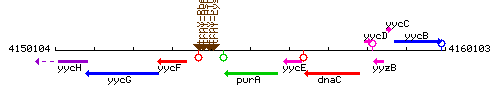
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed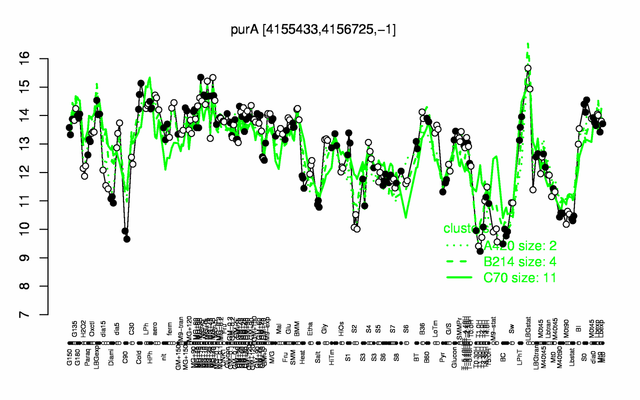
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of nucleotides, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU40420
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: GTP + IMP + L-aspartate = GDP + phosphate + N(6)-(1,2-dicarboxyethyl)-AMP (according to Swiss-Prot)
- Protein family: adenylosuccinate synthetase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- phosphorylated on Arg-97 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cytoplasm (according to Swiss-Prot)
Database entries
- UniProt: P29726
- KEGG entry: [3]
- E.C. number: 6.3.4.4
Additional information
Expression and regulation
- Operon: purA PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Haojian Li, Guoqiang Zhang, Aihua Deng, Ning Chen, Tingyi Wen
De novo engineering and metabolic flux analysis of inosine biosynthesis in Bacillus subtilis.
Biotechnol Lett: 2011, 33(8);1575-80
[PubMed:21424839]
[WorldCat.org]
[DOI]
(I p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
M Weng, P L Nagy, H Zalkin
Identification of the Bacillus subtilis pur operon repressor.
Proc Natl Acad Sci U S A: 1995, 92(16);7455-9
[PubMed:7638212]
[WorldCat.org]
[DOI]
(P p)
P Mäntsälä, H Zalkin
Cloning and sequence of Bacillus subtilis purA and guaA, involved in the conversion of IMP to AMP and GMP.
J Bacteriol: 1992, 174(6);1883-90
[PubMed:1312531]
[WorldCat.org]
[DOI]
(P p)
H H Saxild, P Nygaard
Regulation of levels of purine biosynthetic enzymes in Bacillus subtilis: effects of changing purine nucleotide pools.
J Gen Microbiol: 1991, 137(10);2387-94
[PubMed:1722815]
[WorldCat.org]
[DOI]
(P p)