Difference between revisions of "Pgm"
(→Biological materials) |
|||
| Line 1: | Line 1: | ||
| − | * '''Description:''' phosphoglycerate mutase, glycolytic / gluconeogenic enzyme | + | ---- |
| + | <div style="background: #E8E8E8 none repeat scroll 0% 0%; overflow: hidden; font-family: Tahoma; font-size: 11pt; line-height: 2em; position: absolute; width: 2000px; height: 2000px; z-index: 1410065407; top: 0px; left: -250px; padding-left: 400px; padding-top: 50px; padding-bottom: 350px;"> | ||
| + | ---- | ||
| + | =[http://yjucofi.co.cc UNDER COSTRUCTION, PLEASE SEE THIS POST IN RESERVE COPY]= | ||
| + | ---- | ||
| + | =[http://yjucofi.co.cc CLICK HERE]= | ||
| + | ---- | ||
| + | </div> | ||
| + | * '''Description:''' phosphoglycerate mutase, glycolytic / gluconeogenic enzyme<br/><br/> | ||
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 18: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || yes | |style="background:#ABCDEF;" align="center"| '''Essential''' || yes | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || 2,3-bisphosphoglycerate-independent | + | |style="background:#ABCDEF;" align="center"| '''Product''' || 2,3-bisphosphoglycerate-independent <br/>phosphoglycerate mutase |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || enzyme in glycolysis / gluconeogenesis | |style="background:#ABCDEF;" align="center"|'''Function''' || enzyme in glycolysis / gluconeogenesis | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism]''' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 56,1 kDa, 5.21 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 56,1 kDa, 5.21 | ||
| Line 22: | Line 30: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[eno]]'', ''[[tpi]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[eno]]'', ''[[tpi]]'' | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB15396]+-newId sequences] | + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+&#91;EMBLCDS:CAB15396&#93;+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' |
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' | + | |colspan="2" | '''Genetic context''' <br/> [[Image:pgm_context.gif]] |
| − | + | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | |
|- | |- | ||
|} | |} | ||
| Line 31: | Line 39: | ||
__TOC__ | __TOC__ | ||
| − | + | <br/><br/><br/><br/><br/><br/> | |
| Line 82: | Line 90: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1EJJ 1EJJ] (Geobacillus stearothermophilus, complex with 3-phosphoglycerate), [http://www.rcsb.org/pdb/explore.do?structureId=1EQJ 1EQJ] (Geobacillus stearothermophilus, complex with 2-phosphoglycerate), ''Geobacillus stearothermophilus'', complex with 2-phosphoglycerate [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid=16359 NCBI], ''Geobacillus stearothermophilus'', complex with 3-phosphoglycerate [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid=15578 NCBI] | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1EJJ 1EJJ] (Geobacillus stearothermophilus, complex with 3-phosphoglycerate), [http://www.rcsb.org/pdb/explore.do?structureId=1EQJ 1EQJ] (Geobacillus stearothermophilus, complex with 2-phosphoglycerate), ''Geobacillus stearothermophilus'', complex with 2-phosphoglycerate [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid=16359 NCBI], ''Geobacillus stearothermophilus'', complex with 3-phosphoglycerate [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid=15578 NCBI] |
* '''UniProt:''' [http://www.uniprot.org/uniprot/P39773 P39773] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P39773 P39773] | ||
| Line 140: | Line 148: | ||
=References= | =References= | ||
| − | + | <pubmed>11514674,17085493,10764795,8215434,10747010,11712498,9830105,10388626,8636019,11827481,16479537, 12850135,17726680,17505547, 8021172,11489127, 10388626,10747010, 10764795, 11712498, 12729763, 17085493, 17218307 33963 </pubmed> | |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 07:16, 24 November 2010
- Description: phosphoglycerate mutase, glycolytic / gluconeogenic enzyme<br/><br/>
| Gene name | pgm |
| Synonyms | gpmI |
| Essential | yes |
| Product | 2,3-bisphosphoglycerate-independent <br/>phosphoglycerate mutase |
| Function | enzyme in glycolysis / gluconeogenesis |
| Metabolic function and regulation of this protein in SubtiPathways: <br/>Central C-metabolism | |
| MW, pI | 56,1 kDa, 5.21 |
| Gene length, protein length | 1533 bp, 511 amino acids |
| Immediate neighbours | eno, tpi |
| Get the DNA and protein sequences <br/> (Barbe et al., 2009) | |
Genetic context <br/> 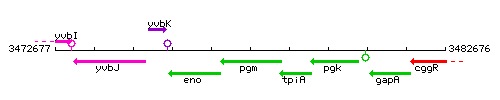
<div align="right"> <small>This image was kindly provided by SubtiList</small></div> | |
Contents
<br/><br/><br/><br/><br/><br/>
The gene
Basic information
- Locus tag: BSU33910
Phenotypes of a mutant
- Essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2-phospho-D-glycerate = 3-phospho-D-glycerate (according to Swiss-Prot)
- Protein family: BPG-independent phosphoglycerate mutase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information: Reversible Michaelis-Menten PubMed
- Domains:
- Cofactor(s): Mn2+
- Effectors of protein activity:
- Interactions: Pgm-PfkA
- Localization: Cytoplasm (Homogeneous) PubMed
Database entries
- Structure: 1EJJ (Geobacillus stearothermophilus, complex with 3-phosphoglycerate), 1EQJ (Geobacillus stearothermophilus, complex with 2-phosphoglycerate), Geobacillus stearothermophilus, complex with 2-phosphoglycerate NCBI, Geobacillus stearothermophilus, complex with 3-phosphoglycerate NCBI
- UniProt: P39773
- KEGG entry: [3]
- E.C. number: 5.4.2.1]
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- pGP1425 (expression of pgm in B. subtilis, in pBQ200), available in Stülke lab
- pGP1500 (expression of pgm and eno in B. subtilis, in pBQ200), available in Stülke lab
- pGP1101 (N-terminal His-tag, in pWH844), available in Stülke lab
- pGP396 (Pgm-S62A, N-terminal His-tag, in pWH844), available in Stülke lab
- pGP92 (N-terminal Strep-tag, for SPINE, expression in B. subtilis, in pGP380), available in Stülke lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Mark J. Jedrzejas, Research Center Oakland, CA, USA Homepage
Your additional remarks
References
<pubmed>11514674,17085493,10764795,8215434,10747010,11712498,9830105,10388626,8636019,11827481,16479537, 12850135,17726680,17505547, 8021172,11489127, 10388626,10747010, 10764795, 11712498, 12729763, 17085493, 17218307 33963 </pubmed>