Difference between revisions of "LmrB"
(→References) |
|||
| Line 140: | Line 140: | ||
=References= | =References= | ||
| − | + | <pubmed>15317768,17483215,12499232, 21815947 </pubmed> | |
| − | |||
| − | |||
| − | |||
| − | <pubmed>15317768,17483215,12499232, </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 16:07, 13 July 2013
- Description: lincomycin-resistance protein (multidrug resistance pump)
| Gene name | lmrB |
| Synonyms | yccA |
| Essential | no |
| Product | lincomycin-resistance protein |
| Function | resistance to lincomycin |
| Gene expression levels in SubtiExpress: lmrB | |
| MW, pI | 51 kDa, 9.703 |
| Gene length, protein length | 1437 bp, 479 aa |
| Immediate neighbours | ycbU, lmrA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 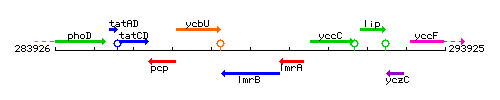
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed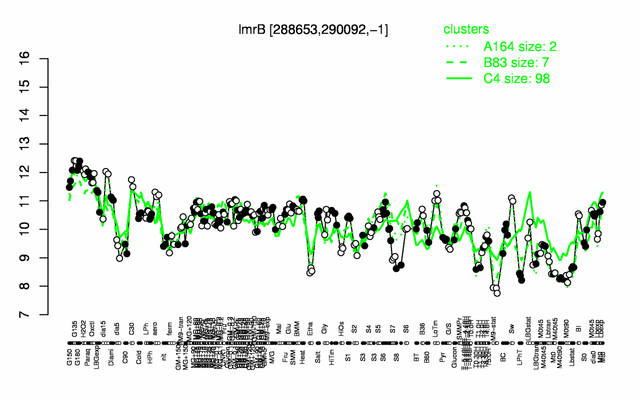
| |
Contents
Categories containing this gene/protein
transporters/ other, resistance against toxins/ antibiotics, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU02670
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: EmrB family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: O35018
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Martin Lehnik-Habrink, Marc Schaffer, Ulrike Mäder, Christine Diethmaier, Christina Herzberg, Jörg Stülke
RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y.
Mol Microbiol: 2011, 81(6);1459-73
[PubMed:21815947]
[WorldCat.org]
[DOI]
(I p)
Kazutake Hirooka, Satoshi Kunikane, Hiroshi Matsuoka, Ken-Ichi Yoshida, Kanako Kumamoto, Shigeo Tojo, Yasutaro Fujita
Dual regulation of the Bacillus subtilis regulon comprising the lmrAB and yxaGH operons and yxaF gene by two transcriptional repressors, LmrA and YxaF, in response to flavonoids.
J Bacteriol: 2007, 189(14);5170-82
[PubMed:17483215]
[WorldCat.org]
[DOI]
(P p)
Ken-Ichi Yoshida, Yo-Hei Ohki, Makiko Murata, Masaki Kinehara, Hiroshi Matsuoka, Takenori Satomura, Reiko Ohki, Miyuki Kumano, Kunio Yamane, Yasutaro Fujita
Bacillus subtilis LmrA is a repressor of the lmrAB and yxaGH operons: identification of its binding site and functional analysis of lmrB and yxaGH.
J Bacteriol: 2004, 186(17);5640-8
[PubMed:15317768]
[WorldCat.org]
[DOI]
(P p)
Miyuki Kumano, Masaya Fujita, Kouji Nakamura, Makiko Murata, Reiko Ohki, Kunio Yamane
Lincomycin resistance mutations in two regions immediately downstream of the -10 region of lmr promoter cause overexpression of a putative multidrug efflux pump in Bacillus subtilis mutants.
Antimicrob Agents Chemother: 2003, 47(1);432-5
[PubMed:12499232]
[WorldCat.org]
[DOI]
(P p)