Difference between revisions of "CitB"
(→Biological materials) |
|||
| Line 39: | Line 39: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | + | <br/><br/> | |
| − | |||
| − | |||
| − | |||
| − | |||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| Line 146: | Line 142: | ||
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant:''' GP683 (erm) | + | * '''Mutant:''' |
| + | ** GP683 (erm), available in [[Jörg Stülke]]'s lab | ||
| + | ** GP1441 (spc), available in [[Jörg Stülke]]'s lab | ||
** 1A999 ( ''citB''::''spec''), {{PubMed| }}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A999&Search=1A999 BGSC] | ** 1A999 ( ''citB''::''spec''), {{PubMed| }}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A999&Search=1A999 BGSC] | ||
* '''Expression vector:''' | * '''Expression vector:''' | ||
| − | * '''lacZ fusion:''' pGP700 (in [[pAC5]]), available in [[Stülke]] lab | + | * '''lacZ fusion:''' |
| + | ** pGP700 (in [[pAC5]]), available in [[Jörg Stülke]]'s lab | ||
* '''GFP fusion:''' | * '''GFP fusion:''' | ||
| − | * '''two-hybrid system:''' ''B. pertussis'' adenylate cyclase-based bacterial two hybrid system ([[BACTH]]), available in [[Stülke]] lab | + | * '''two-hybrid system:''' ''B. pertussis'' adenylate cyclase-based bacterial two hybrid system ([[BACTH]]), available in [[Jörg Stülke]]'s lab |
| − | * '''Antibody:''' available in [[Linc Sonenshein]] lab | + | * '''Antibody:''' available in [[Linc Sonenshein]]'s lab |
| − | * '''FLAG-tag construct:''' GP1144 (spc, based on [[pGP1331]]) | + | * '''FLAG-tag construct:''' |
| + | ** GP1144 (spc, based on [[pGP1331]]), available in [[Jörg Stülke]]'s lab | ||
| + | ** GP1145 (kan), available in [[Jörg Stülke]]'s lab | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
Revision as of 17:13, 30 October 2012
- Description: trigger enzyme: aconitase and RNA binding protein
| Gene name | citB |
| Synonyms | |
| Essential | no |
| Product | trigger enzyme: aconitate hydratase (aconitase) |
| Function | TCA cycle |
| Gene expression levels in SubtiExpress: citB | |
| Interactions involving this protein in SubtInteract: CitB | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 99 kDa, 4.903 |
| Gene length, protein length | 2727 bp, 909 aa |
| Immediate neighbours | sspO, yneN |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 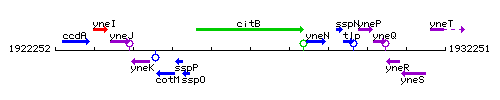
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed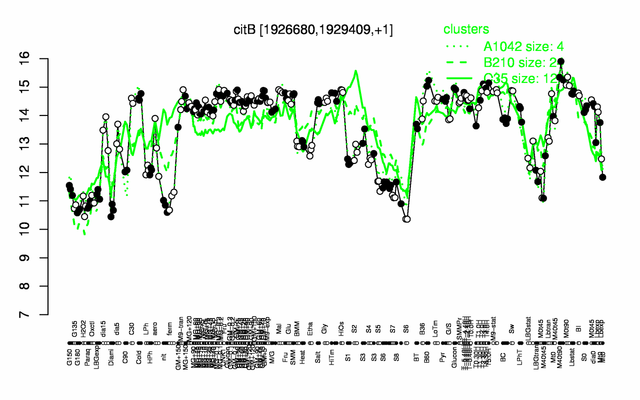
| |
Contents
Categories containing this gene/protein
carbon core metabolism, trigger enzyme, RNA binding regulators
This gene is a member of the following regulons
CcpC regulon, CodY regulon, FsrA regulon
The CitB regulon: feuA-feuB-feuC-ybbA
The gene
Basic information
- Locus tag: BSU18000
Phenotypes of a mutant
glutamate auxotrophy and a defect in sporulation PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Citrate = isocitrate (according to Swiss-Prot)
- Binding to iron responsive elements (IRE RNA) in the absence of the FeS cluster PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): FeS cluster
- Effectors of protein activity:
Database entries
- Structure: 1L5J (E. coli)
- UniProt: P09339
- KEGG entry: [3]
- E.C. number: 4.2.1.3
Additional information
- B. subtilis aconitase is both an enzyme and an RNA binding protein (moonlighting protein) PubMed
- extensive information on the structure and enzymatic properties of CitB can be found at Proteopedia
Expression and regulation
- Regulation:
- repressed during growth in the presence of branched chain amino acids (CodY) PubMed
- repressed in the presence of glucose and glutamate (CcpC) PubMed
- expressed upon transition into the stationary phase (AbrB) PubMed, indirect negative regulation by AbrB PubMed
- repressed by glucose (3.7-fold) (CcpA) PubMed
- repression by glucose + arginine (CcpC) PubMed
- less expressed under conditions of extreme iron limitation (FsrA) PubMed
- part of the iron sparing response (FsrA) PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- GP683 (erm), available in Jörg Stülke's lab
- GP1441 (spc), available in Jörg Stülke's lab
- 1A999 ( citB::spec), PubMed, available at BGSC
- Expression vector:
- lacZ fusion:
- pGP700 (in pAC5), available in Jörg Stülke's lab
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- Antibody: available in Linc Sonenshein's lab
- FLAG-tag construct:
- GP1144 (spc, based on pGP1331), available in Jörg Stülke's lab
- GP1145 (kan), available in Jörg Stülke's lab
Labs working on this gene/protein
Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
Reviews
Karl Volz
The functional duality of iron regulatory protein 1.
Curr Opin Struct Biol: 2008, 18(1);106-11
[PubMed:18261896]
[WorldCat.org]
[DOI]
(P p)
Fabian M Commichau, Jörg Stülke
Trigger enzymes: bifunctional proteins active in metabolism and in controlling gene expression.
Mol Microbiol: 2008, 67(4);692-702
[PubMed:18086213]
[WorldCat.org]
[DOI]
(P p)
Patricia J Kiley, Helmut Beinert
The role of Fe-S proteins in sensing and regulation in bacteria.
Curr Opin Microbiol: 2003, 6(2);181-5
[PubMed:12732309]
[WorldCat.org]
[DOI]
(P p)
R L Switzer
Non-redox roles for iron-sulfur clusters in enzymes.
Biofactors: 1989, 2(2);77-86
[PubMed:2696478]
[WorldCat.org]
(P p)
Original publications