SnaA
- Description: sulphur N-acetylase, required for the conversion of S-methyl-cysteine to cysteine
| Gene name | ytmI |
| Synonyms | |
| Essential | no |
| Product | sulphur N-acetylase |
| Function | utilization of S-methyl-cysteine |
| Gene expression levels in SubtiExpress: snaA | |
| Metabolic function and regulation of this protein in SubtiPathways: Riboflavin / FAD | |
| MW, pI | 20 kDa, 5.508 |
| Gene length, protein length | 534 bp, 178 aa |
| Immediate neighbours | tcyJ, ascR |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed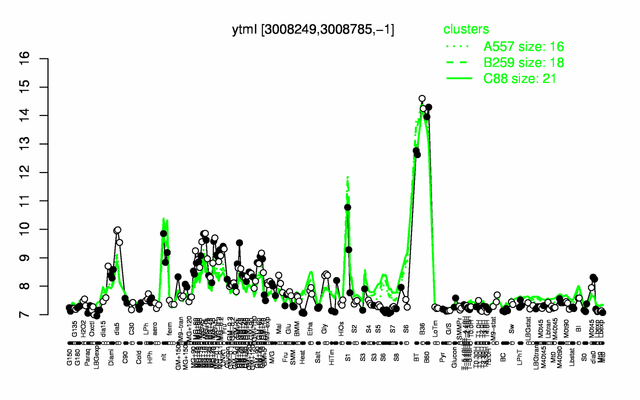
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29390
Phenotypes of a mutant
- reduced growth with S-methyl cysteine PubMed
- the snaA snaB double mutant does not grow with S-methyl cysteine PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- N-acetylation of the amino group of S-methyl cysteine PubMed
- Protein family: acetyltransferase family (according to Swiss-Prot)
- Paralogous protein(s): SnaB
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O34350
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Che-Man Chan, Antoine Danchin, Philippe Marlière, Agnieszka Sekowska
Paralogous metabolism: S-alkyl-cysteine degradation in Bacillus subtilis.
Environ Microbiol: 2014, 16(1);101-17
[PubMed:23944997]
[WorldCat.org]
[DOI]
(I p)
Pierre Burguière, Juliette Fert, Isabelle Guillouard, Sandrine Auger, Antoine Danchin, Isabelle Martin-Verstraete
Regulation of the Bacillus subtilis ytmI operon, involved in sulfur metabolism.
J Bacteriol: 2005, 187(17);6019-30
[PubMed:16109943]
[WorldCat.org]
[DOI]
(P p)
Kyle N Erwin, Shunji Nakano, Peter Zuber
Sulfate-dependent repression of genes that function in organosulfur metabolism in Bacillus subtilis requires Spx.
J Bacteriol: 2005, 187(12);4042-9
[PubMed:15937167]
[WorldCat.org]
[DOI]
(P p)
Irina M Solovieva, Rimma A Kreneva, Lubov Errais Lopes, Daniel A Perumov
The riboflavin kinase encoding gene ribR of Bacillus subtilis is a part of a 10 kb operon, which is negatively regulated by the yrzC gene product.
FEMS Microbiol Lett: 2005, 243(1);51-8
[PubMed:15668000]
[WorldCat.org]
[DOI]
(P p)
Jean-Yves Coppée, Sandrine Auger, Evelyne Turlin, Agnieszka Sekowska, Jean-Pierre Le Caer, Valérie Labas, Valérie Vagner, Antoine Danchin, Isabelle Martin-Verstraete
Sulfur-limitation-regulated proteins in Bacillus subtilis: a two-dimensional gel electrophoresis study.
Microbiology (Reading): 2001, 147(Pt 6);1631-1640
[PubMed:11390694]
[WorldCat.org]
[DOI]
(P p)