Difference between revisions of "YydG"
(→Original publications) |
|||
| Line 143: | Line 143: | ||
<pubmed> 23106164</pubmed> | <pubmed> 23106164</pubmed> | ||
==Original publications== | ==Original publications== | ||
| − | + | <pubmed>,17921301,12850135 17921301, 21815947</pubmed> | |
| − | |||
| − | |||
| − | |||
| − | <pubmed>,17921301,12850135 17921301, </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 16:41, 13 July 2013
- Description: oxidoreductase, modification of YydF
| Gene name | yydG |
| Synonyms | |
| Essential | no |
| Product | oxidoreductase |
| Function | control of LiaR-LiaS activity |
| Gene expression levels in SubtiExpress: yydG | |
| Interactions involving this protein in SubtInteract: YydG | |
| MW, pI | 37 kDa, 5.865 |
| Gene length, protein length | 957 bp, 319 aa |
| Immediate neighbours | yydH, yydF |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 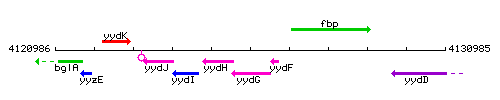
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed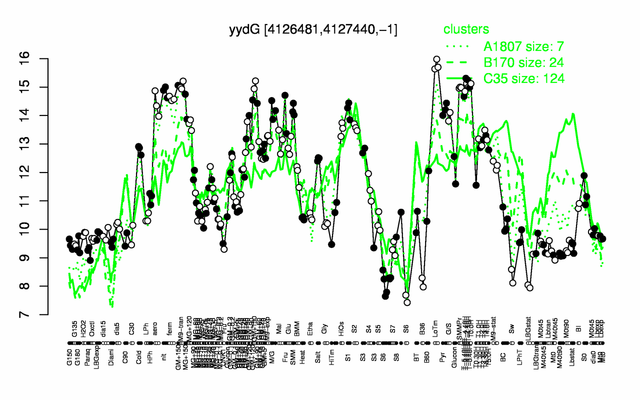
| |
Contents
Categories containing this gene/protein
transcription factors and their control
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU40170
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: Q45595
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Susanne Gebhard
ABC transporters of antimicrobial peptides in Firmicutes bacteria - phylogeny, function and regulation.
Mol Microbiol: 2012, 86(6);1295-317
[PubMed:23106164]
[WorldCat.org]
[DOI]
(I p)
Original publications
Martin Lehnik-Habrink, Marc Schaffer, Ulrike Mäder, Christine Diethmaier, Christina Herzberg, Jörg Stülke
RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y.
Mol Microbiol: 2011, 81(6);1459-73
[PubMed:21815947]
[WorldCat.org]
[DOI]
(I p)
Bronwyn G Butcher, Yi-Pin Lin, John D Helmann
The yydFGHIJ operon of Bacillus subtilis encodes a peptide that induces the LiaRS two-component system.
J Bacteriol: 2007, 189(23);8616-25
[PubMed:17921301]
[WorldCat.org]
[DOI]
(I p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)