Difference between revisions of "YydG"
| Line 78: | Line 78: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| − | * '''Localization:''' | + | * '''[[Localization]]:''' |
=== Database entries === | === Database entries === | ||
| Line 108: | Line 108: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** the amount of the mRNA is substantially decreased upon depletion of [[Rny|RNase Y]] {{PubMed|21815947}} | ||
=Biological materials = | =Biological materials = | ||
| Line 128: | Line 129: | ||
=References= | =References= | ||
| − | + | <big>''Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J'' </big> | |
| + | <big>'''RNA processing in ''Bacillus subtilis'': identification of targets of the essential RNase Y.''' </big> | ||
| + | <big>Mol Microbiol. 2011 81(6): 1459-1473. </big> | ||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/21815947 PubMed:21815947] | ||
<pubmed>,17921301,12850135 17921301, </pubmed> | <pubmed>,17921301,12850135 17921301, </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 14:32, 20 November 2011
- Description: oxidoreductase
| Gene name | yydG |
| Synonyms | |
| Essential | no |
| Product | oxidoreductase |
| Function | control of LiaR-LiaS activity |
| MW, pI | 37 kDa, 5.865 |
| Gene length, protein length | 957 bp, 319 aa |
| Immediate neighbours | yydH, yydF |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 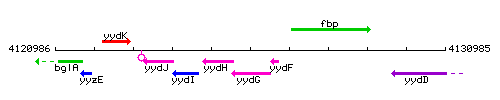
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
transcription factors and their control
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU40170
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: Q45595
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol Microbiol. 2011 81(6): 1459-1473. PubMed:21815947