Difference between revisions of "YxxG"
| Line 45: | Line 45: | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
{{SubtiWiki regulon|[[DegU regulon]]}}, | {{SubtiWiki regulon|[[DegU regulon]]}}, | ||
| + | {{SubtiWiki regulon|[[WapA regulon]]}}, | ||
{{SubtiWiki regulon|[[YvrHb regulon]]}} | {{SubtiWiki regulon|[[YvrHb regulon]]}} | ||
| Line 63: | Line 64: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| Line 82: | Line 81: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' phosphorylation on (Tyr-102 OR Ser-105) [http://www.ncbi.nlm.nih.gov/sites/entrez/17218307 PubMed] | * '''Modification:''' phosphorylation on (Tyr-102 OR Ser-105) [http://www.ncbi.nlm.nih.gov/sites/entrez/17218307 PubMed] | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 113: | Line 112: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yxxG_4023054_4023482_-1 yxxG] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yxxG_4023054_4023482_-1 yxxG] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|23199363}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 121: | Line 120: | ||
** [[DegU]]-P: transcription repression {{PubMed|9537385}} | ** [[DegU]]-P: transcription repression {{PubMed|9537385}} | ||
** activated by [[YvrHb]] [http://www.ncbi.nlm.nih.gov/sites/entrez/16306698 PubMed] | ** activated by [[YvrHb]] [http://www.ncbi.nlm.nih.gov/sites/entrez/16306698 PubMed] | ||
| + | ** [[WalR]]: transcription repression {{PubMed|23199363}} | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| Line 144: | Line 144: | ||
=References= | =References= | ||
| − | <pubmed>16672620, 20815827,17218307,16306698 9537385 23572593</pubmed> | + | <pubmed>16672620, 20815827,17218307,16306698 9537385 23572593 23199363</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 11:36, 29 December 2013
- Description: immunity protein, protects the cell against the toxic activity of WapA
| Gene name | yxxG |
| Synonyms | wapI |
| Essential | no |
| Product | immunity protein |
| Function | intercellular competition |
| Gene expression levels in SubtiExpress: yxxG | |
| Interactions involving this protein in SubtInteract: YxxG | |
| MW, pI | 16 kDa, 4.424 |
| Gene length, protein length | 426 bp, 142 aa |
| Immediate neighbours | yxiF, wapA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 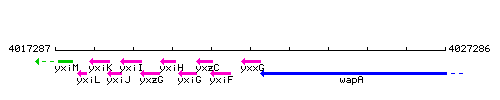
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed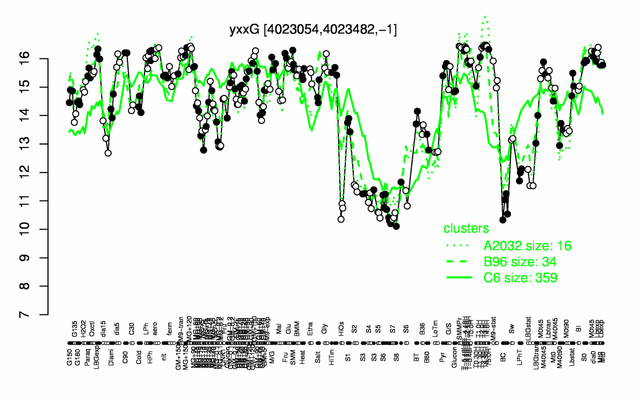
| |
Contents
Categories containing this gene/protein
toxins, antitoxins and immunity against toxins, phosphoproteins,
This gene is a member of the following regulons
DegU regulon, WapA regulon, YvrHb regulon
The gene
Basic information
- Locus tag: BSU39220
Phenotypes of a mutant
- severe growth defect PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification: phosphorylation on (Tyr-102 OR Ser-105) PubMed
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: Q07836
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Sanna Koskiniemi, James G Lamoureux, Kiel C Nikolakakis, Claire t'Kint de Roodenbeke, Michael D Kaplan, David A Low, Christopher S Hayes
Rhs proteins from diverse bacteria mediate intercellular competition.
Proc Natl Acad Sci U S A: 2013, 110(17);7032-7
[PubMed:23572593]
[WorldCat.org]
[DOI]
(I p)
Letal I Salzberg, Leagh Powell, Karsten Hokamp, Eric Botella, David Noone, Kevin M Devine
The WalRK (YycFG) and σ(I) RsgI regulators cooperate to control CwlO and LytE expression in exponentially growing and stressed Bacillus subtilis cells.
Mol Microbiol: 2013, 87(1);180-95
[PubMed:23199363]
[WorldCat.org]
[DOI]
(I p)
Taryn B Kiley, Nicola R Stanley-Wall
Post-translational control of Bacillus subtilis biofilm formation mediated by tyrosine phosphorylation.
Mol Microbiol: 2010, 78(4);947-63
[PubMed:20815827]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Juliane Ollinger, Kyung-Bok Song, Haike Antelmann, Michael Hecker, John D Helmann
Role of the Fur regulon in iron transport in Bacillus subtilis.
J Bacteriol: 2006, 188(10);3664-73
[PubMed:16672620]
[WorldCat.org]
[DOI]
(P p)
Masakuni Serizawa, Keisuke Kodama, Hiroki Yamamoto, Kazuo Kobayashi, Naotake Ogasawara, Junichi Sekiguchi
Functional analysis of the YvrGHb two-component system of Bacillus subtilis: identification of the regulated genes by DNA microarray and northern blot analyses.
Biosci Biotechnol Biochem: 2005, 69(11);2155-69
[PubMed:16306698]
[WorldCat.org]
[DOI]
(P p)
V Dartois, M Débarbouillé, F Kunst, G Rapoport
Characterization of a novel member of the DegS-DegU regulon affected by salt stress in Bacillus subtilis.
J Bacteriol: 1998, 180(7);1855-61
[PubMed:9537385]
[WorldCat.org]
[DOI]
(P p)