Difference between revisions of "YxaL"
| Line 61: | Line 61: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU39940&redirect=T BSU39940] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yxaJL.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yxaJL.html] | ||
| Line 99: | Line 100: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU39940&redirect=T BSU39940] | ||
* '''Structure:''' | * '''Structure:''' | ||
Revision as of 16:14, 2 April 2014
- Description: similar to Ser/Thr kinase, increases the processivity of the PcrA helicase
| Gene name | yxaL |
| Synonyms | yxaK |
| Essential | no |
| Product | unknown |
| Function | unknown |
| Gene expression levels in SubtiExpress: yxaL | |
| Interactions involving this protein in SubtInteract: YxaL | |
| MW, pI | 43 kDa, 6.379 |
| Gene length, protein length | 1230 bp, 410 aa |
| Immediate neighbours | yxaM, yxaJ |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 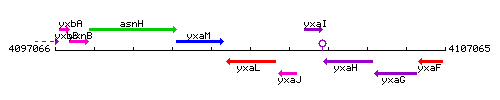
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed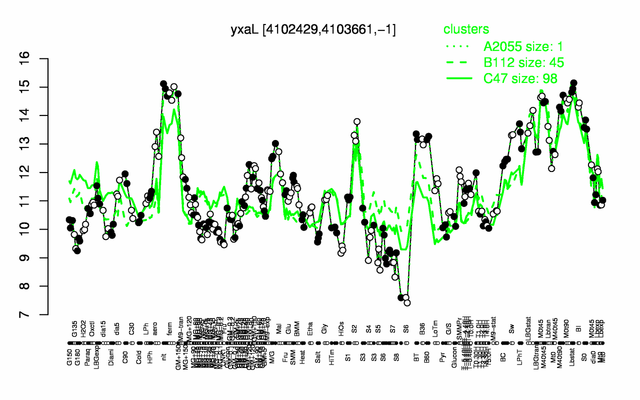
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU39940
Phenotypes of a mutant
Database entries
- BsubCyc: BSU39940
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: extracellular (signal peptide), major constituent of the secretome PubMed
Database entries
- BsubCyc: BSU39940
- Structure:
- UniProt: P42111
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- 'lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Mark Albano, Wiep Klaas Smits, Linh T Y Ho, Barbara Kraigher, Ines Mandic-Mulec, Oscar P Kuipers, David Dubnau
The Rok protein of Bacillus subtilis represses genes for cell surface and extracellular functions.
J Bacteriol: 2005, 187(6);2010-9
[PubMed:15743949]
[WorldCat.org]
[DOI]
(P p)
M-F Noirot-Gros, P Soultanas, D B Wigley, S D Ehrlich, P Noirot, M-A Petit
The beta-propeller protein YxaL increases the processivity of the PcrA helicase.
Mol Genet Genomics: 2002, 267(3);391-400
[PubMed:12073041]
[WorldCat.org]
[DOI]
(P p)
Ken-Ichi Yoshida, Izumi Ishio, Eishi Nagakawa, Yoshiyuki Yamamoto, Mami Yamamoto, Yasutaro Fujita
Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome.
Microbiology (Reading): 2000, 146 ( Pt 3);573-579
[PubMed:10746760]
[WorldCat.org]
[DOI]
(P p)