Difference between revisions of "YvrHb"
(→Extended information on the protein) |
|||
| Line 88: | Line 88: | ||
* '''Effectors of protein activity:''' phosphorylation likely affects DNA-binding activity | * '''Effectors of protein activity:''' phosphorylation likely affects DNA-binding activity | ||
| − | * '''Interactions:''' [[YvrG]]-[[YvrHb]] [http://www.ncbi.nlm.nih.gov/sites/entrez/16306698 PubMed] | + | * '''[[SubtInteract|Interactions]]:''' |
| + | ** [[YvrG]]-[[YvrHb]] [http://www.ncbi.nlm.nih.gov/sites/entrez/16306698 PubMed] | ||
| − | * '''Localization:''' cytoplasm (according to Swiss-Prot) | + | * '''[[Localization]]:''' cytoplasm (according to Swiss-Prot) |
=== Database entries === | === Database entries === | ||
Revision as of 15:58, 10 August 2011
- Description: two-component response regulator
| Gene name | yvrHb |
| Synonyms | yvrH |
| Essential | no |
| Product | two-component response regulator |
| Function | regulation cell surface maintenance |
| Interactions involving this protein in SubtInteract: YvrHB | |
| MW, pI | 26.6 kDa, 5.23 |
| Gene length, protein length | 714 bp, 237 amino acids |
| Immediate neighbours | yvrG, yvrHa |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 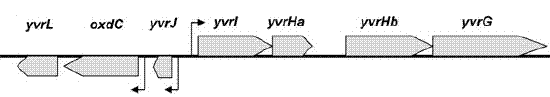
| |
Contents
Categories containing this gene/protein
transcription factors and their control, phosphoproteins
This gene is a member of the following regulons
The YvrHb regulon
The gene
Basic information
- Locus tag:
Phenotypes of a mutant
unusual autolysis and higher susceptibility to cell envelope antibiotics (bacitracin, aztreonam, cefepime, fosfomycin) PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: no entry
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcription activator PubMed
- Protein family: OmpR family
- Paralogous protein(s):
The YvrHb regulon
subject to positive control: dltA-dltB-dltC-dltD-dltE, sigX-rsiX, sunA, sunT-bdbA-yolJ-bdbB, wapA-yxxG, wprA, yvrI-yvrHa PubMed
subject to negative control: lytA-lytB-lytC PubMed
Extended information on the protein
- Kinetic information:
- Domains:
- Cofactor(s):
- Effectors of protein activity: phosphorylation likely affects DNA-binding activity
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- Swiss prot entry: P94504
- KEGG entry: [1]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Letal I Salzberg, John D Helmann
Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition.
J Bacteriol: 2008, 190(23);7797-807
[PubMed:18820022]
[WorldCat.org]
[DOI]
(I p)
Masakuni Serizawa, Keisuke Kodama, Hiroki Yamamoto, Kazuo Kobayashi, Naotake Ogasawara, Junichi Sekiguchi
Functional analysis of the YvrGHb two-component system of Bacillus subtilis: identification of the regulated genes by DNA microarray and northern blot analyses.
Biosci Biotechnol Biochem: 2005, 69(11);2155-69
[PubMed:16306698]
[WorldCat.org]
[DOI]
(P p)
C Fabret, V A Feher, J A Hoch
Two-component signal transduction in Bacillus subtilis: how one organism sees its world.
J Bacteriol: 1999, 181(7);1975-83
[PubMed:10094672]
[WorldCat.org]
[DOI]
(P p)