Difference between revisions of "YvgJ"
| Line 22: | Line 22: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yvsG]]'', ''[[yvgK]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yvsG]]'', ''[[yvgK]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU33360 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU33360 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU33360 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU33360 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU33360 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU33360 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:yvgJ_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:yvgJ_context.gif]] | ||
Revision as of 11:15, 14 May 2013
- Description: lipoteichoic acid synthesis primase
| Gene name | yvgJ |
| Synonyms | yvsF |
| Essential | no |
| Product | lipoteichoic acid synthesis primase |
| Function | biosynthesis of lipoteichoic acid |
| Gene expression levels in SubtiExpress: yvgJ | |
| MW, pI | 70 kDa, 5.31 |
| Gene length, protein length | 1851 bp, 617 aa |
| Immediate neighbours | yvsG, yvgK |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 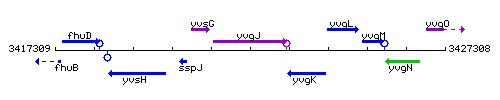
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed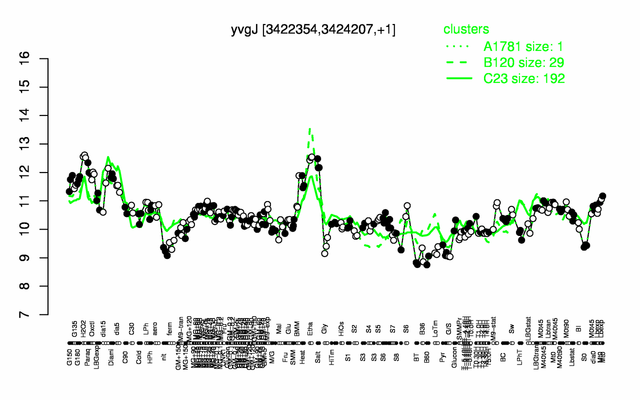
| |
Contents
Categories containing this gene/protein
cell wall synthesis, biosynthesis of cell wall components, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU33360
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: LTA synthase family (according to Swiss-Prot)
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: O32206
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Additional reviews: PubMed
Nathalie T Reichmann, Angelika Gründling
Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes.
FEMS Microbiol Lett: 2011, 319(2);97-105
[PubMed:21388439]
[WorldCat.org]
[DOI]
(I p)
Original publications
Michihiro Hashimoto, Takahiro Seki, Satoshi Matsuoka, Hiroshi Hara, Kei Asai, Yoshito Sadaie, Kouji Matsumoto
Induction of extracytoplasmic function sigma factors in Bacillus subtilis cells with defects in lipoteichoic acid synthesis.
Microbiology (Reading): 2013, 159(Pt 1);23-35
[PubMed:23103977]
[WorldCat.org]
[DOI]
(I p)
Mirka E Wörmann, Rebecca M Corrigan, Peter J Simpson, Steve J Matthews, Angelika Gründling
Enzymatic activities and functional interdependencies of Bacillus subtilis lipoteichoic acid synthesis enzymes.
Mol Microbiol: 2011, 79(3);566-83
[PubMed:21255105]
[WorldCat.org]
[DOI]
(I p)
Kathrin Schirner, Jon Marles-Wright, Richard J Lewis, Jeff Errington
Distinct and essential morphogenic functions for wall- and lipo-teichoic acids in Bacillus subtilis.
EMBO J: 2009, 28(7);830-42
[PubMed:19229300]
[WorldCat.org]
[DOI]
(I p)