Difference between revisions of "YveA"
| Line 27: | Line 27: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| − | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yveA_3539165_3540727_1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:yveA_expression.png|500px]] | + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yveA_3539165_3540727_1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:yveA_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU34470]] |
|- | |- | ||
|} | |} | ||
Revision as of 14:38, 16 May 2013
- Description: aspartate/ glutamate transporter
| Gene name | yveA |
| Synonyms | |
| Essential | no |
| Product | aspartate/ glutamate transporter |
| Function | aspartate uptake |
| Gene expression levels in SubtiExpress: yveA | |
| MW, pI | 56 kDa, 9.675 |
| Gene length, protein length | 1560 bp, 520 aa |
| Immediate neighbours | levB, yvdT |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 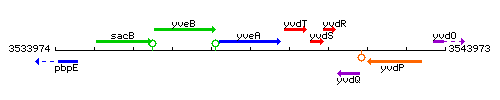
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed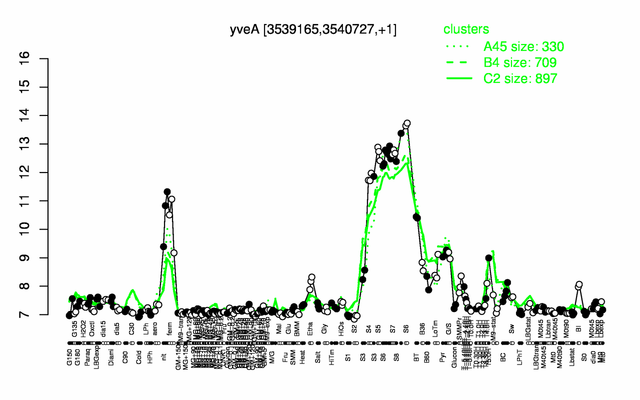
| |
Contents
Categories containing this gene/protein
transporters/ other, biosynthesis/ acquisition of amino acids, utilization of amino acids, sporulation proteins, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU34470
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): YbeC
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: O07002
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- SacY: binding to a RNA switch (transcriptional antitermination) PubMed
- DegU-P: transcription activation PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Stephanie T Wang, Barbara Setlow, Erin M Conlon, Jessica L Lyon, Daisuke Imamura, Tsutomu Sato, Peter Setlow, Richard Losick, Patrick Eichenberger
The forespore line of gene expression in Bacillus subtilis.
J Mol Biol: 2006, 358(1);16-37
[PubMed:16497325]
[WorldCat.org]
[DOI]
(P p)
Graciela Lorca, Brit Winnen, Milton H Saier
Identification of the L-aspartate transporter in Bacillus subtilis.
J Bacteriol: 2003, 185(10);3218-22
[PubMed:12730183]
[WorldCat.org]
[DOI]
(P p)
Y Pereira, M F Petit-Glatron, R Chambert
yveB, Encoding endolevanase LevB, is part of the sacB-yveB-yveA levansucrase tricistronic operon in Bacillus subtilis.
Microbiology (Reading): 2001, 147(Pt 12);3413-9
[PubMed:11739774]
[WorldCat.org]
[DOI]
(P p)
S Aymerich, M Steinmetz
Cloning and preliminary characterization of the sacS locus from Bacillus subtilis which controls the regulation of the exoenzyme levansucrase.
Mol Gen Genet: 1987, 208(1-2);114-20
[PubMed:3039303]
[WorldCat.org]
[DOI]
(P p)
H Shimotsu, D J Henner
Modulation of Bacillus subtilis levansucrase gene expression by sucrose and regulation of the steady-state mRNA level by sacU and sacQ genes.
J Bacteriol: 1986, 168(1);380-8
[PubMed:2428811]
[WorldCat.org]
[DOI]
(P p)