Difference between revisions of "YtdI"
| Line 37: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 117: | Line 113: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| + | ** [[RoxS]]: inhibition of translation and initiation of RNA degradation by [[rny|RNase Y]] and [[rnc|RNase III]] {{PubMed|25643072}} | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** the mRNA base-pairs with the [[RoxS]] [[sRNA]] {{PubMed|25643072}} | ||
| + | ** half-life of the mRNA: 8.4 min, in a ''[[roxS]]'' mutant 15 min, in a ''[[rny]]'' mutant 24 min {{PubMed|25643072}} | ||
** the mRNA is substantially stabilized upon depletion of [[Rny|RNase Y]] {{PubMed|21815947}} | ** the mRNA is substantially stabilized upon depletion of [[Rny|RNase Y]] {{PubMed|21815947}} | ||
| + | ** two pathways of mRNA degradation: (i) by [[rny|RNase Y]], (ii) by combined action of [[RoxS]] and [[rnc|RNase III]] {{PubMed|25643072}} | ||
** number of protein molecules per cell (minimal medium with glucose and ammonium): 714 {{PubMed|24696501}} | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 714 {{PubMed|24696501}} | ||
** number of protein molecules per cell (complex medium with amino acids, without glucose): 1384 {{PubMed|24696501}} | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 1384 {{PubMed|24696501}} | ||
| Line 142: | Line 142: | ||
=References= | =References= | ||
| − | <pubmed>16511117 | + | <pubmed>16511117 25643072 21815947</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:45, 4 February 2015
- Description: inorganic polyphosphate/ATP-NAD kinase
| Gene name | ytdI |
| Synonyms | ppnKB |
| Essential | no |
| Product | inorganic polyphosphate/ATP-NAD kinase |
| Function | generation of NADP from NAD |
| Gene expression levels in SubtiExpress: ytdI | |
| Metabolic function and regulation of this protein in SubtiPathways: YtdI | |
| MW, pI | 30 kDa, 6.49 |
| Gene length, protein length | 801 bp, 267 aa |
| Immediate neighbours | sppA, ytcJ |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 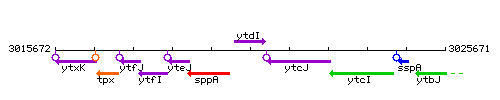
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29540
Phenotypes of a mutant
Database entries
- BsubCyc: BSU29540
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + NAD+ = ADP + NADP+ (according to Swiss-Prot)
- Protein family: NAD kinase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU29540
- UniProt: O34934
- KEGG entry: [2]
- E.C. number: 2.7.1.23
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
- the mRNA base-pairs with the RoxS sRNA PubMed
- half-life of the mRNA: 8.4 min, in a roxS mutant 15 min, in a rny mutant 24 min PubMed
- the mRNA is substantially stabilized upon depletion of RNase Y PubMed
- two pathways of mRNA degradation: (i) by RNase Y, (ii) by combined action of RoxS and RNase III PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 714 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 1384 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Sylvain Durand, Frédérique Braun, Efthimia Lioliou, Cédric Romilly, Anne-Catherine Helfer, Laurianne Kuhn, Noé Quittot, Pierre Nicolas, Pascale Romby, Ciarán Condon
A nitric oxide regulated small RNA controls expression of genes involved in redox homeostasis in Bacillus subtilis.
PLoS Genet: 2015, 11(2);e1004957
[PubMed:25643072]
[WorldCat.org]
[DOI]
(I e)
Martin Lehnik-Habrink, Marc Schaffer, Ulrike Mäder, Christine Diethmaier, Christina Herzberg, Jörg Stülke
RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y.
Mol Microbiol: 2011, 81(6);1459-73
[PubMed:21815947]
[WorldCat.org]
[DOI]
(I p)
Vaheh Oganesyan, Candice Huang, Paul D Adams, Jaru Jancarik, Hisao A Yokota, Rosalind Kim, Sung-Hou Kim
Structure of a NAD kinase from Thermotoga maritima at 2.3 A resolution.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2005, 61(Pt 7);640-6
[PubMed:16511117]
[WorldCat.org]
[DOI]
(I p)