Difference between revisions of "YtdI"
(→References) |
|||
| Line 56: | Line 56: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU29540&redirect=T BSU29540] | ||
* '''DBTBS entry:''' no entry | * '''DBTBS entry:''' no entry | ||
| Line 91: | Line 92: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU29540&redirect=T BSU29540] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1YT5 1YT5] (from ''Thermotoga maritima'') [http://www.ncbi.nlm.nih.gov/sites/entrez/16511117 PubMed] | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1YT5 1YT5] (from ''Thermotoga maritima'') [http://www.ncbi.nlm.nih.gov/sites/entrez/16511117 PubMed] | ||
Revision as of 14:31, 2 April 2014
- Description: inorganic polyphosphate/ATP-NAD kinase
| Gene name | ytdI |
| Synonyms | ppnKB |
| Essential | no |
| Product | inorganic polyphosphate/ATP-NAD kinase |
| Function | generation of NADP from NAD |
| Gene expression levels in SubtiExpress: ytdI | |
| MW, pI | 30 kDa, 6.49 |
| Gene length, protein length | 801 bp, 267 aa |
| Immediate neighbours | sppA, ytcJ |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 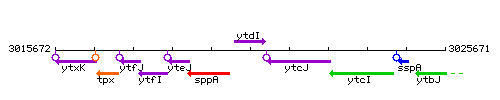
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed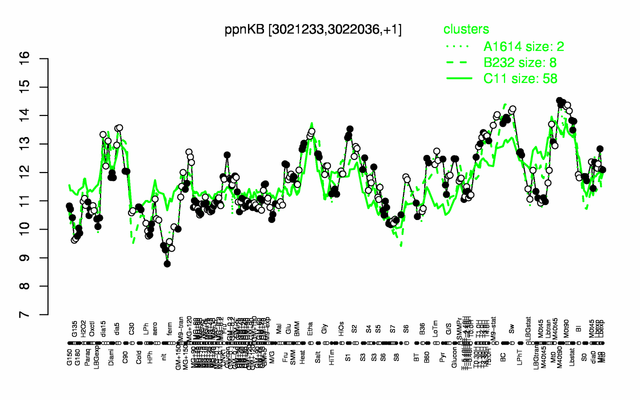
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29540
Phenotypes of a mutant
Database entries
- BsubCyc: BSU29540
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + NAD+ = ADP + NADP+ (according to Swiss-Prot)
- Protein family: NAD kinase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU29540
- UniProt: O34934
- KEGG entry: [2]
- E.C. number: 2.7.1.23
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Martin Lehnik-Habrink, Marc Schaffer, Ulrike Mäder, Christine Diethmaier, Christina Herzberg, Jörg Stülke
RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y.
Mol Microbiol: 2011, 81(6);1459-73
[PubMed:21815947]
[WorldCat.org]
[DOI]
(I p)
Vaheh Oganesyan, Candice Huang, Paul D Adams, Jaru Jancarik, Hisao A Yokota, Rosalind Kim, Sung-Hou Kim
Structure of a NAD kinase from Thermotoga maritima at 2.3 A resolution.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2005, 61(Pt 7);640-6
[PubMed:16511117]
[WorldCat.org]
[DOI]
(I p)