Difference between revisions of "YtaG"
| Line 42: | Line 42: | ||
{{SubtiWiki category|[[biosynthesis of cofactors]]}}, | {{SubtiWiki category|[[biosynthesis of cofactors]]}}, | ||
{{SubtiWiki category|[[essential genes]]}}, | {{SubtiWiki category|[[essential genes]]}}, | ||
| − | {{SubtiWiki category|[[phosphoproteins]]}} | + | {{SubtiWiki category|[[phosphoproteins]]}}, |
| + | {{SubtiWiki category|[[sporulation/ other]]}} | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
Revision as of 08:24, 22 April 2014
- Description: dephospho-CoA kinase
| Gene name | ytaG |
| Synonyms | coaE |
| Essential | yes PubMed |
| Product | dephospho-CoA kinase |
| Function | synthesis of coenzyme A |
| Gene expression levels in SubtiExpress: ytaG | |
| Metabolic function and regulation of this protein in SubtiPathways: ytaG | |
| MW, pI | 21 kDa, 4.858 |
| Gene length, protein length | 591 bp, 197 aa |
| Immediate neighbours | ytbE, ytaF |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 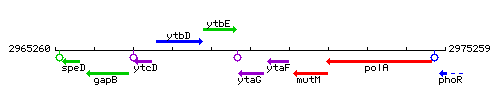
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed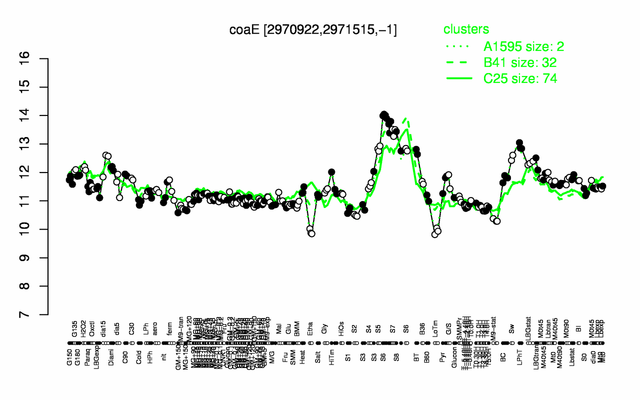
| |
Contents
Categories containing this gene/protein
biosynthesis of cofactors, essential genes, phosphoproteins, sporulation/ other
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29060
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU29060
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + 3'-dephospho-CoA = ADP + CoA (according to Swiss-Prot)
- Protein family: coaE family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylated on ser/ thr/ tyr PubMed
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- BsubCyc: BSU29060
- UniProt: O34932
- KEGG entry: [3]
- E.C. number: 2.7.1.24
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Alain Lévine, Françoise Vannier, Cédric Absalon, Lauriane Kuhn, Peter Jackson, Elaine Scrivener, Valérie Labas, Joëlle Vinh, Patrick Courtney, Jérôme Garin, Simone J Séror
Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes.
Proteomics: 2006, 6(7);2157-73
[PubMed:16493705]
[WorldCat.org]
[DOI]
(P p)
Nicholas O'Toole, João A R G Barbosa, Yunge Li, Li-Wei Hung, Allan Matte, Miroslaw Cygler
Crystal structure of a trimeric form of dephosphocoenzyme A kinase from Escherichia coli.
Protein Sci: 2003, 12(2);327-36
[PubMed:12538896]
[WorldCat.org]
[DOI]
(P p)
P Mishra, P K Park, D G Drueckhammer
Identification of yacE (coaE) as the structural gene for dephosphocoenzyme A kinase in Escherichia coli K-12.
J Bacteriol: 2001, 183(9);2774-8
[PubMed:11292795]
[WorldCat.org]
[DOI]
(P p)
Alia Lapidus, Nathalie Galleron, Alexei Sorokin, S Dusko Ehrlich
Sequencing and functional annotation of the Bacillus subtilis genes in the 200 kb rrnB-dnaB region.
Microbiology (Reading): 1997, 143 ( Pt 11);3431-3441
[PubMed:9387221]
[WorldCat.org]
[DOI]
(P p)