Difference between revisions of "YtaB"
| Line 35: | Line 35: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 63: | Line 59: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 96: | Line 89: | ||
* '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU30930&redirect=T BSU30930] | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU30930&redirect=T BSU30930] | ||
| − | * '''Structure:''' | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore/explore.do?structureId=4RYR 4RYR] (the protein from B. cereus, 56% identity, 89% similarity) {{PubMed|25635100}} |
* '''UniProt:''' [http://www.uniprot.org/uniprot/O34694 O34694] | * '''UniProt:''' [http://www.uniprot.org/uniprot/O34694 O34694] | ||
| Line 140: | Line 133: | ||
=References= | =References= | ||
| − | <pubmed>15805528,9387221 | + | <pubmed>15805528,9387221 25635100 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 16:27, 16 February 2015
- Description: general stress protein, survival of ethanol and salt stresses
| Gene name | ytaB |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | survival of ethanol and salt stresses |
| Gene expression levels in SubtiExpress: ytaB | |
| MW, pI | 17 kDa, 9.73 |
| Gene length, protein length | 465 bp, 155 aa |
| Immediate neighbours | cotI, glgP |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 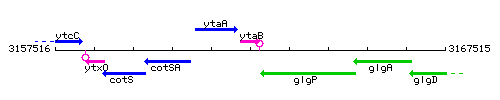
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed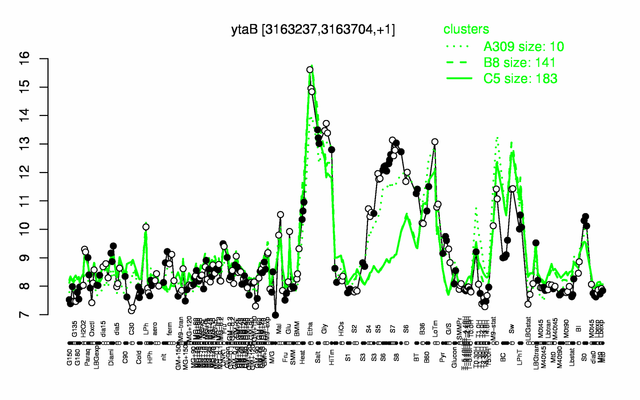
| |
Contents
Categories containing this gene/protein
general stress proteins (controlled by SigB)
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU30930
Phenotypes of a mutant
Database entries
- BsubCyc: BSU30930
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- BsubCyc: BSU30930
- UniProt: O34694
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Youzhong Guo, Ravi C Kalathur, Qun Liu, Brian Kloss, Renato Bruni, Christopher Ginter, Edda Kloppmann, Burkhard Rost, Wayne A Hendrickson
Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science: 2015, 347(6221);551-5
[PubMed:25635100]
[WorldCat.org]
[DOI]
(I p)
Dirk Höper, Uwe Völker, Michael Hecker
Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis.
J Bacteriol: 2005, 187(8);2810-26
[PubMed:15805528]
[WorldCat.org]
[DOI]
(P p)
Alia Lapidus, Nathalie Galleron, Alexei Sorokin, S Dusko Ehrlich
Sequencing and functional annotation of the Bacillus subtilis genes in the 200 kb rrnB-dnaB region.
Microbiology (Reading): 1997, 143 ( Pt 11);3431-3441
[PubMed:9387221]
[WorldCat.org]
[DOI]
(P p)