Difference between revisions of "YsbA"
(→References) |
|||
| Line 137: | Line 137: | ||
=References= | =References= | ||
| − | <pubmed> 23175651 | + | <pubmed> 23175651 21815947 15659658 17452642</pubmed> |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 16:34, 13 July 2013
- Description: putative anti-holin
| Gene name | ysbA |
| Synonyms | lrgA |
| Essential | no |
| Product | putative anti-holin |
| Function | unknown |
| Gene expression levels in SubtiExpress: ysbA | |
| MW, pI | 15 kDa, 9.825 |
| Gene length, protein length | 438 bp, 146 aa |
| Immediate neighbours | ysbB, lytT |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 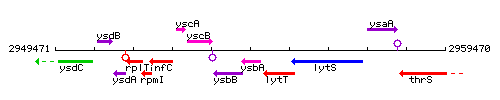
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed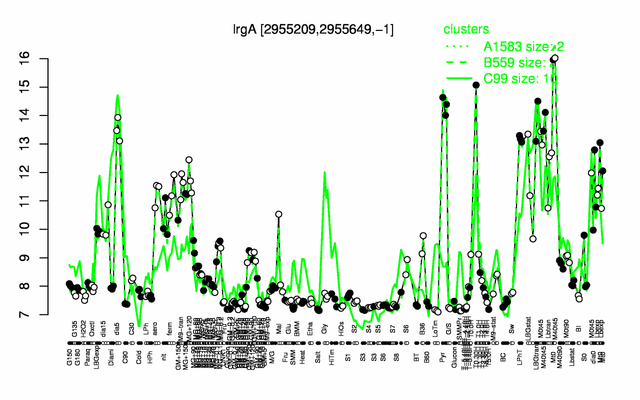
| |
Contents
Categories containing this gene/protein
membrane proteins, proteins of unknown function
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU28910
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: LrgA subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P94515
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- GP1554 (ysbA-ysbB::spc), available in Jörg Stülke's lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Martin Lehnik-Habrink, Leonie Rempeters, Ákos T Kovács, Christoph Wrede, Claudia Baierlein, Heike Krebber, Oscar P Kuipers, Jörg Stülke
DEAD-Box RNA helicases in Bacillus subtilis have multiple functions and act independently from each other.
J Bacteriol: 2013, 195(3);534-44
[PubMed:23175651]
[WorldCat.org]
[DOI]
(I p)
Martin Lehnik-Habrink, Marc Schaffer, Ulrike Mäder, Christine Diethmaier, Christina Herzberg, Jörg Stülke
RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y.
Mol Microbiol: 2011, 81(6);1459-73
[PubMed:21815947]
[WorldCat.org]
[DOI]
(I p)
Kelly C Rice, Ethan E Mann, Jennifer L Endres, Elizabeth C Weiss, James E Cassat, Mark S Smeltzer, Kenneth W Bayles
The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus.
Proc Natl Acad Sci U S A: 2007, 104(19);8113-8
[PubMed:17452642]
[WorldCat.org]
[DOI]
(P p)
Kelly C Rice, Jeremy B Nelson, Toni G Patton, Soo-Jin Yang, Kenneth W Bayles
Acetic acid induces expression of the Staphylococcus aureus cidABC and lrgAB murein hydrolase regulator operons.
J Bacteriol: 2005, 187(3);813-21
[PubMed:15659658]
[WorldCat.org]
[DOI]
(P p)