Difference between revisions of "YraA"
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || degradation of damaged thiol-containing proteins | |style="background:#ABCDEF;" align="center"|'''Function''' || degradation of damaged thiol-containing proteins | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU27020 yraA] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 18 kDa, 4.729 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 18 kDa, 4.729 | ||
Revision as of 14:15, 7 August 2012
- Description: general stress protein, degradation of damaged thiol-containing proteins
| Gene name | yraA |
| Synonyms | |
| Essential | no |
| Product | cysteine proteinase |
| Function | degradation of damaged thiol-containing proteins |
| Gene expression levels in SubtiExpress: yraA | |
| MW, pI | 18 kDa, 4.729 |
| Gene length, protein length | 507 bp, 169 aa |
| Immediate neighbours | adhA, sacC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 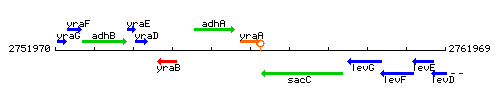
This image was kindly provided by SubtiList
| |
| Expression at a glance PubMed 500px | |
Contents
Categories containing this gene/protein
proteolysis, general stress proteins (controlled by SigB), resistance against oxidative and electrophile stress
This gene is a member of the following regulons
AdhR regulon, SigB regulon, SigM regulon
The gene
Basic information
- Locus tag: BSU27020
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: peptidase C56 family (according to Swiss-Prot)
- Paralogous protein(s): YfkM
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: O06006
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Thi Thu Huyen Nguyen, Warawan Eiamphungporn, Ulrike Mäder, Manuel Liebeke, Michael Lalk, Michael Hecker, John D Helmann, Haike Antelmann
Genome-wide responses to carbonyl electrophiles in Bacillus subtilis: control of the thiol-dependent formaldehyde dehydrogenase AdhA and cysteine proteinase YraA by the MerR-family regulator YraB (AdhR).
Mol Microbiol: 2009, 71(4);876-94
[PubMed:19170879]
[WorldCat.org]
[DOI]
(I p)
Adrian J Jervis, Penny D Thackray, Chris W Houston, Malcolm J Horsburgh, Anne Moir
SigM-responsive genes of Bacillus subtilis and their promoters.
J Bacteriol: 2007, 189(12);4534-8
[PubMed:17434969]
[WorldCat.org]
[DOI]
(P p)
Dirk Höper, Uwe Völker, Michael Hecker
Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis.
J Bacteriol: 2005, 187(8);2810-26
[PubMed:15805528]
[WorldCat.org]
[DOI]
(P p)