Difference between revisions of "YqiQ"
| Line 16: | Line 16: | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU24120 yqiQ] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU24120 yqiQ] | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/ | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/subtipathways/search.php?enzyme=yqiQ yqiQ]''' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 32 kDa, 5.279 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 32 kDa, 5.279 | ||
Revision as of 11:10, 7 January 2014
- Description: methylisocitrate lyase
| Gene name | yqiQ |
| Synonyms | prpB |
| Essential | no |
| Product | methylisocitrate lyase |
| Function | mother cell metabolism |
| Gene expression levels in SubtiExpress: yqiQ | |
| Metabolic function and regulation of this protein in SubtiPathways: yqiQ | |
| MW, pI | 32 kDa, 5.279 |
| Gene length, protein length | 903 bp, 301 aa |
| Immediate neighbours | yqzF, mmgE |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 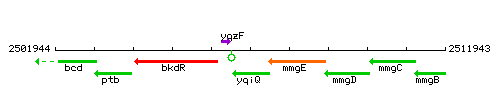
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed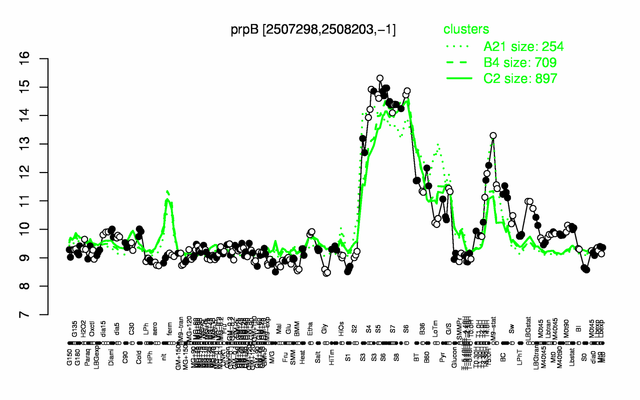
| |
Contents
Categories containing this gene/protein
sporulation proteins, membrane proteins, poorly characterized/ putative enzymes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU24120
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: (2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate = pyruvate + succinate (according to Swiss-Prot)
- Protein family: Methylisocitrate lyase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
Database entries
- UniProt: P54528
- KEGG entry: [3]
- E.C. number: 4.1.3.30
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Leif Steil, Mónica Serrano, Adriano O Henriques, Uwe Völker
Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis.
Microbiology (Reading): 2005, 151(Pt 2);399-420
[PubMed:15699190]
[WorldCat.org]
[DOI]
(P p)
Clemens Grimm, Andreas Evers, Matthias Brock, Claudia Maerker, Gerhard Klebe, Wolfgang Buckel, Klaus Reuter
Crystal structure of 2-methylisocitrate lyase (PrpB) from Escherichia coli and modelling of its ligand bound active centre.
J Mol Biol: 2003, 328(3);609-21
[PubMed:12706720]
[WorldCat.org]
[DOI]
(P p)
E M Bryan, B W Beall, C P Moran
A sigma E dependent operon subject to catabolite repression during sporulation in Bacillus subtilis.
J Bacteriol: 1996, 178(16);4778-86
[PubMed:8759838]
[WorldCat.org]
[DOI]
(P p)