Difference between revisions of "YqgP"
| Line 6: | Line 6: | ||
|''yqgP'' | |''yqgP'' | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' glpG'' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
| Line 93: | Line 93: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| + | ** forms dimers {{PubMed|22921757}} | ||
| − | * '''Localization:''' membrane | + | * '''[[Localization]]:''' |
| + | ** cell membrane {{PubMed|22921757}} | ||
=== Database entries === | === Database entries === | ||
| Line 143: | Line 145: | ||
=References= | =References= | ||
| − | <pubmed>19421188,15050034,, </pubmed> | + | <pubmed>19421188,15050034,22921757, </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:20, 28 August 2012
- Description: intramembrane protease
| Gene name | yqgP |
| Synonyms | glpG |
| Essential | no |
| Product | intramembrane protease |
| Function | cell division or sugar metabolism |
| Gene expression levels in SubtiExpress: yqgP | |
| MW, pI | 56 kDa, 5.507 |
| Gene length, protein length | 1521 bp, 507 aa |
| Immediate neighbours | yqgQ, yqgO |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 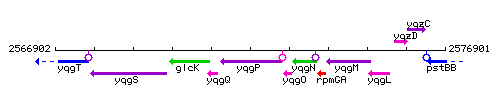
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
proteolysis, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU24870
Phenotypes of a mutant
filamentous phenotype PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Cleaves type-1 transmembrane domains using a catalytic dyad composed of serine and histidine that are contributed by different transmembrane domains (according to Swiss-Prot)
- Protein family: peptidase S54 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- forms dimers PubMed
- Localization:
- cell membrane PubMed
Database entries
- Structure:
- UniProt: P54493
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Padmapriya Sampathkumar, Michelle W Mak, Sarah J Fischer-Witholt, Emmanuel Guigard, Cyril M Kay, M Joanne Lemieux
Oligomeric state study of prokaryotic rhomboid proteases.
Biochim Biophys Acta: 2012, 1818(12);3090-7
[PubMed:22921757]
[WorldCat.org]
[DOI]
(P p)
Sinisa Urban
Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms.
Nat Rev Microbiol: 2009, 7(6);411-23
[PubMed:19421188]
[WorldCat.org]
[DOI]
(I p)
Lili R Mesak, Felix M Mesak, Michael K Dahl
Expression of a novel gene, gluP, is essential for normal Bacillus subtilis cell division and contributes to glucose export.
BMC Microbiol: 2004, 4;13
[PubMed:15050034]
[WorldCat.org]
[DOI]
(I e)