Difference between revisions of "YmdB"
(→References) |
|||
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || control of bistable gene expression | |style="background:#ABCDEF;" align="center"|'''Function''' || control of bistable gene expression | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU16970 ymdB] |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 29,1 kDa, 6.50 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 29,1 kDa, 6.50 | ||
| Line 22: | Line 22: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[rny]]'', ''[[spoVS]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[rny]]'', ''[[spoVS]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU16970 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU16970 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU16970 Advanced_DNA] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:ymdB_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:ymdB_context.gif]] | ||
Revision as of 13:53, 13 May 2013
- Description: phosphodiesterase, controls bistable gene expression
| Gene name | ymdB |
| Synonyms | |
| Essential | no |
| Product | phosphodiesterase |
| Function | control of bistable gene expression |
| Gene expression levels in SubtiExpress: ymdB | |
| MW, pI | 29,1 kDa, 6.50 |
| Gene length, protein length | 792 bp, 264 amino acids |
| Immediate neighbours | rny, spoVS |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 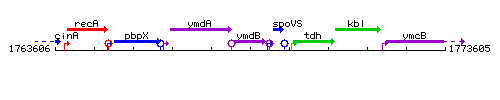
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed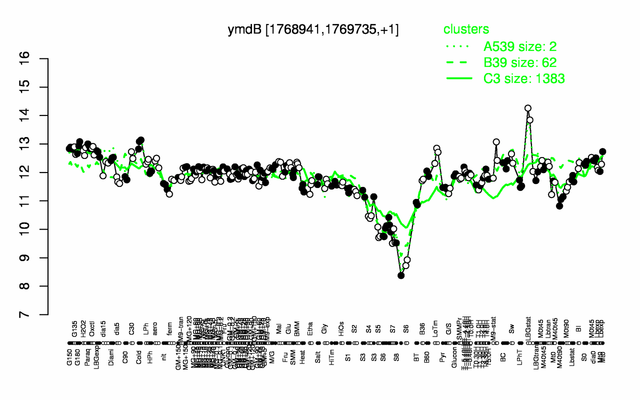
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU16970
Phenotypes of a mutant
- strong overexpression of hag PubMed
- defective in biofilm formation PubMed
- the phenotypes of the ymdB mutant can be suppressed by overexpression of slrR PubMed
- inactivation of ymdB restores beta-lactam resistance in a sigM mutant PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: phosphatase activity toward phosphoenolpyruvate and phosphodiesterase activity toward 2',3'-cAMP (the Deinococcus ortholog) PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: O31775
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation: constitutive
- Regulatory mechanism:
- Additional information: there is a terminator between rny and ymdB, most transcripts terminate there PubMed
Biological materials
- Mutant:
- Expression vector:
- for expression/ purification from B. subtilis with N-terminal Strep-tag, for SPINE, in pGP380: pGP1041, available in Stülke lab
- for expression/ purification from B. subtilis with C-terminal Strep-tag, for SPINE, in pGP382: pGP1919, available in Stülke lab
- for expression/ purification from E. coli with N-terminal His-tag, in pWH844: pGP1040, available in Stülke lab
- for expression/ purification from E. coli with N-terminal Strep-tag, in pGP172: pGP1917, available in Stülke lab
- GP970 (ymdB-Strep (cat)), purification from B. subtilis, for SPINE, available in Stülke lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
Yun Luo, John D Helmann
Analysis of the role of Bacillus subtilis σ(M) in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis.
Mol Microbiol: 2012, 83(3);623-39
[PubMed:22211522]
[WorldCat.org]
[DOI]
(I p)
Eric R Pozsgai, Kris M Blair, Daniel B Kearns
Modified mariner transposons for random inducible-expression insertions and transcriptional reporter fusion insertions in Bacillus subtilis.
Appl Environ Microbiol: 2012, 78(3);778-85
[PubMed:22113911]
[WorldCat.org]
[DOI]
(I p)
Christine Diethmaier, Nico Pietack, Katrin Gunka, Christoph Wrede, Martin Lehnik-Habrink, Christina Herzberg, Sebastian Hübner, Jörg Stülke
A novel factor controlling bistability in Bacillus subtilis: the YmdB protein affects flagellin expression and biofilm formation.
J Bacteriol: 2011, 193(21);5997-6007
[PubMed:21856853]
[WorldCat.org]
[DOI]
(I p)
Functional and structural analysis of orthologs in other organisms
Jason Zemansky, Benjamin C Kline, Joshua J Woodward, Jess H Leber, Hélène Marquis, Daniel A Portnoy
Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype.
J Bacteriol: 2009, 191(12);3950-64
[PubMed:19376879]
[WorldCat.org]
[DOI]
(I p)
Dong Hae Shin, Michael Proudfoot, Hyo Jin Lim, In-Kyu Choi, Hisao Yokota, Alexander F Yakunin, Rosalind Kim, Sung-Hou Kim
Structural and enzymatic characterization of DR1281: A calcineurin-like phosphoesterase from Deinococcus radiodurans.
Proteins: 2008, 70(3);1000-9
[PubMed:17847097]
[WorldCat.org]
[DOI]
(I p)