Difference between revisions of "YkoV"
(→References) |
(→References) |
||
| Line 138: | Line 138: | ||
=References= | =References= | ||
== Reviews == | == Reviews == | ||
| − | <pubmed> 11591342 11719239 </pubmed> | + | <pubmed> 11591342 11719239 17938628 </pubmed> |
== Original publications == | == Original publications == | ||
<pubmed>16497325, 11483577, 23691176 12215643 </pubmed> | <pubmed>16497325, 11483577, 23691176 12215643 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 10:55, 23 May 2013
- Description: confers dry-heat resistance to dormant spores PubMed
| Gene name | ykoV |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | confers dry-heat resistance to dormant spores |
| Gene expression levels in SubtiExpress: ykoV | |
| MW, pI | 34 kDa, 7.213 |
| Gene length, protein length | 933 bp, 311 aa |
| Immediate neighbours | ykoU, ykoW |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 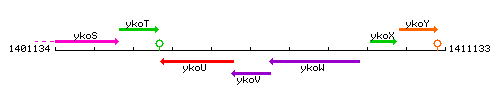
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed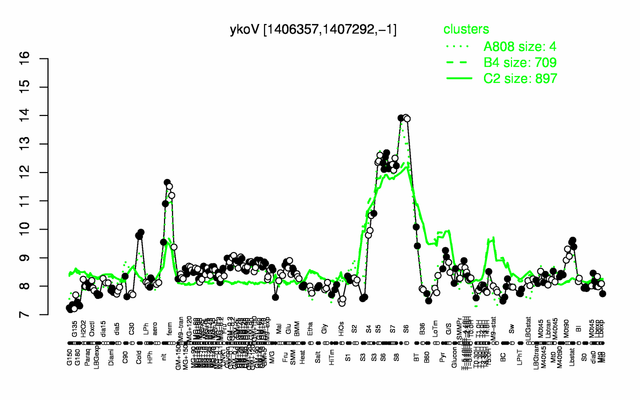
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU13410
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- associates with the nucleoid during germination PubMed
Database entries
- Structure:
- UniProt: O34859
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Stewart Shuman, Michael S Glickman
Bacterial DNA repair by non-homologous end joining.
Nat Rev Microbiol: 2007, 5(11);852-61
[PubMed:17938628]
[WorldCat.org]
[DOI]
(I p)
A J Doherty, S P Jackson
DNA repair: how Ku makes ends meet.
Curr Biol: 2001, 11(22);R920-4
[PubMed:11719239]
[WorldCat.org]
[DOI]
(P p)
J M Jones, M Gellert, W Yang
A Ku bridge over broken DNA.
Structure: 2001, 9(10);881-4
[PubMed:11591342]
[WorldCat.org]
[DOI]
(P p)
Original publications
Miguel de Vega
The minimal Bacillus subtilis nonhomologous end joining repair machinery.
PLoS One: 2013, 8(5);e64232
[PubMed:23691176]
[WorldCat.org]
[DOI]
(I e)
Stephanie T Wang, Barbara Setlow, Erin M Conlon, Jessica L Lyon, Daisuke Imamura, Tsutomu Sato, Peter Setlow, Richard Losick, Patrick Eichenberger
The forespore line of gene expression in Bacillus subtilis.
J Mol Biol: 2006, 358(1);16-37
[PubMed:16497325]
[WorldCat.org]
[DOI]
(P p)
Geoffrey R Weller, Boris Kysela, Rajat Roy, Louise M Tonkin, Elizabeth Scanlan, Marina Della, Susanne Krogh Devine, Jonathan P Day, Adam Wilkinson, Fabrizio d'Adda di Fagagna, Kevin M Devine, Richard P Bowater, Penny A Jeggo, Stephen P Jackson, Aidan J Doherty
Identification of a DNA nonhomologous end-joining complex in bacteria.
Science: 2002, 297(5587);1686-9
[PubMed:12215643]
[WorldCat.org]
[DOI]
(I p)
L Aravind, E V Koonin
Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system.
Genome Res: 2001, 11(8);1365-74
[PubMed:11483577]
[WorldCat.org]
[DOI]
(P p)