Difference between revisions of "YhdR"
| Line 1: | Line 1: | ||
| − | + | ||
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || unknown | |style="background:#ABCDEF;" align="center"|'''Function''' || unknown | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU09570 yhdR] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 43 kDa, 5.007 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 43 kDa, 5.007 | ||
Revision as of 16:19, 6 August 2012
| Gene name | yhdR |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | unknown |
| Gene expression levels in SubtiExpress: yhdR | |
| MW, pI | 43 kDa, 5.007 |
| Gene length, protein length | 1179 bp, 393 aa |
| Immediate neighbours | cueR, yhdS |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 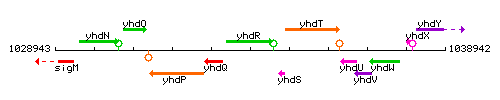
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
poorly characterized/ putative enzymes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU09570
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: L-aspartate + 2-oxoglutarate = oxaloacetate + L-glutamate (according to Swiss-Prot)
- Protein family: class-I pyridoxal-phosphate-dependent aminotransferase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: O07587
- KEGG entry: [2]
- E.C. number: 2.6.1.1
Additional information
The gene is annotated in KEGG as aspartate aminotransferase EC 2.6.1.1. In MetaCyc the protein is marked as “similar to aspartate aminotransferase”. No EC annotation is available in Swiss-Prot. No literature/experimental evidence supporting the annotation is availablavailable. PubMed
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody: