Difference between revisions of "YfhK"
| Line 88: | Line 88: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[yfhK]]-[[yfhL]]-[[yfhM]]'' {{PubMed|10913081}} |
| − | * '''[[Sigma factor]]:''' [[SigB]] [ | + | * '''[[Sigma factor]]:''' [[SigB]] {{PubMed|10913081,15805528}}, [[SigW]] {{PubMed|9987136}} |
| − | * '''Regulation:''' repressed by casamino acids [http://www.ncbi.nlm.nih.gov/pubmed/12107147 PubMed] | + | * '''Regulation:''' |
| + | ** repressed by casamino acids [http://www.ncbi.nlm.nih.gov/pubmed/12107147 PubMed] | ||
| + | ** induced by stress ([[SigB]]) {{PubMed|10913081,15805528}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| Line 118: | Line 120: | ||
=References= | =References= | ||
| − | <pubmed>12107147 15805528, </pubmed> | + | <pubmed>12107147 15805528, 10913081 9987136 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 18:46, 2 September 2009
- Description: general stress protein, similar to cell division inhibitor
| Gene name | yfhK |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | survival of ethonol stress and at low and high temperatures |
| MW, pI | 18 kDa, 9.96 |
| Gene length, protein length | 516 bp, 172 aa |
| Immediate neighbours | yfhJ, yfhL |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 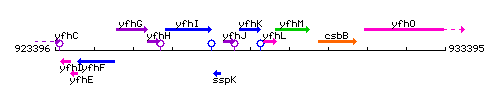
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU08570
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: O31579
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Dirk Höper, Uwe Völker, Michael Hecker
Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis.
J Bacteriol: 2005, 187(8);2810-26
[PubMed:15805528]
[WorldCat.org]
[DOI]
(P p)
Ulrike Mäder, Georg Homuth, Christian Scharf, Knut Büttner, Rüdiger Bode, Michael Hecker
Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability.
J Bacteriol: 2002, 184(15);4288-95
[PubMed:12107147]
[WorldCat.org]
[DOI]
(P p)
H Antelmann, C Scharf, M Hecker
Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis.
J Bacteriol: 2000, 182(16);4478-90
[PubMed:10913081]
[WorldCat.org]
[DOI]
(P p)
X Huang, A Gaballa, M Cao, J D Helmann
Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, sigma W.
Mol Microbiol: 1999, 31(1);361-71
[PubMed:9987136]
[WorldCat.org]
[DOI]
(P p)