YdaE
- Description: lyxose isomerase, general stress protein, survival of ethanol stress and low temperatures
| Gene name | ydaE |
| Synonyms | |
| Essential | no |
| Product | lyxose isomerase |
| Function | survival of ethanol stress and low temperatures |
| Gene expression levels in SubtiExpress: ydaE | |
| MW, pI | 19 kDa, 4.878 |
| Gene length, protein length | 501 bp, 167 aa |
| Immediate neighbours | ydaD, ydaF |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 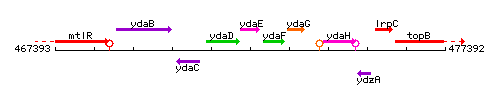
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed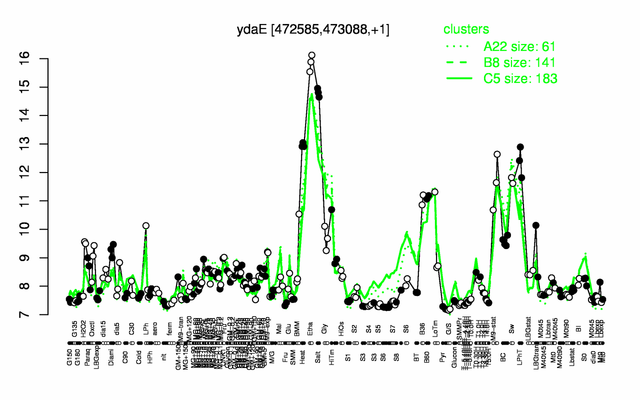
| |
Contents
Categories containing this gene/protein
general stress proteins (controlled by SigB), Utilization of specific carbon sources
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU04200
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: D-lyxose = D-xylulose (according to Swiss-Prot)
- Protein family: D-lyxose ketol-isomerase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P96578
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Imke G de Jong, Jan-Willem Veening, Oscar P Kuipers
Single cell analysis of gene expression patterns during carbon starvation in Bacillus subtilis reveals large phenotypic variation.
Environ Microbiol: 2012, 14(12);3110-21
[PubMed:23033921]
[WorldCat.org]
[DOI]
(I p)
Jon Marles-Wright, Richard J Lewis
The structure of a D-lyxose isomerase from the σB regulon of Bacillus subtilis.
Proteins: 2011, 79(6);2015-9
[PubMed:21520290]
[WorldCat.org]
[DOI]
(I p)
Dirk Höper, Uwe Völker, Michael Hecker
Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis.
J Bacteriol: 2005, 187(8);2810-26
[PubMed:15805528]
[WorldCat.org]
[DOI]
(P p)
Anja Petersohn, Haike Antelmann, Ulf Gerth, Michael Hecker
Identification and transcriptional analysis of new members of the sigmaB regulon in Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 4);869-880
[PubMed:10220166]
[WorldCat.org]
[DOI]
(P p)