Difference between revisions of "YclQ"
| Line 52: | Line 52: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' uptake of the siderophore petrobactin {{PubMed|19955416}} |
* '''Protein family:''' Fe/B12 periplasmic-binding domain (according to Swiss-Prot) | * '''Protein family:''' Fe/B12 periplasmic-binding domain (according to Swiss-Prot) | ||
| Line 88: | Line 88: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[yclN]]-[[yclO]]-[[yclP]]-[[yclQ]]'' {{PubMed|19955416}} |
* '''[[Sigma factor]]:''' | * '''[[Sigma factor]]:''' | ||
| Line 120: | Line 120: | ||
=References= | =References= | ||
| − | <pubmed>10092453,,12354229,18957862 18763711, </pubmed> | + | <pubmed>10092453, 19955416, 12354229,18957862 18763711, </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 10:20, 4 December 2009
- Description: petrobactin (3.4-catecholate siderophore) ABC transporter (binding protein), major component of the secretome
| Gene name | yclQ |
| Synonyms | |
| Essential | no |
| Product | petrobactin ABC transporter (binding protein) |
| Function | iron acquisition |
| MW, pI | 34 kDa, 6.628 |
| Gene length, protein length | 951 bp, 317 aa |
| Immediate neighbours | yclP, ycnB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 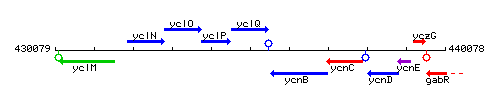
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU03830
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: uptake of the siderophore petrobactin PubMed
- Protein family: Fe/B12 periplasmic-binding domain (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: secreted (according to Swiss-Prot), extracellular (signal peptide) PubMed, membrane PubMed
Database entries
- Structure: 3GFV
- UniProt: P94421
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Anna M Zawadzka, Youngchang Kim, Natalia Maltseva, Rita Nichiporuk, Yao Fan, Andrzej Joachimiak, Kenneth N Raymond
Characterization of a Bacillus subtilis transporter for petrobactin, an anthrax stealth siderophore.
Proc Natl Acad Sci U S A: 2009, 106(51);21854-9
[PubMed:19955416]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Noel Baichoo, Tao Wang, Rick Ye, John D Helmann
Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon.
Mol Microbiol: 2002, 45(6);1613-29
[PubMed:12354229]
[WorldCat.org]
[DOI]
(P p)
Y Quentin, G Fichant, F Denizot
Inventory, assembly and analysis of Bacillus subtilis ABC transport systems.
J Mol Biol: 1999, 287(3);467-84
[PubMed:10092453]
[WorldCat.org]
[DOI]
(P p)