Difference between revisions of "YceE"
| Line 22: | Line 22: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yceD]]'', ''[[yceF]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yceD]]'', ''[[yceF]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU02910 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU02910 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU02910 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU02910 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU02910 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU02910 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:yceE_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:yceE_context.gif]] | ||
Revision as of 09:25, 14 May 2013
- Description: general stress protein, similar to tellurium resistance protein
| Gene name | yceE |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | required for survival of ethanol stress and at low temperatures |
| Gene expression levels in SubtiExpress: yceE | |
| MW, pI | 20 kDa, 4.391 |
| Gene length, protein length | 576 bp, 192 aa |
| Immediate neighbours | yceD, yceF |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 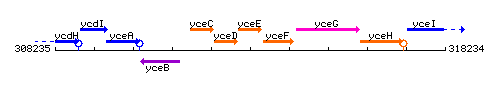
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
general stress proteins (controlled by SigB), cell envelope stress proteins (controlled by SigM, V, W, X, Y), resistance against toxic metals, phosphoproteins
This gene is a member of the following regulons
SigB regulon, SigM regulon, SigW regulon, Sigx regulon
The gene
Basic information
- Locus tag: BSU02910
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: CAPAB/terDEXZ family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- phosphorylated on Arg-10 PubMed
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O34384
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Warawan Eiamphungporn, John D Helmann
Extracytoplasmic function sigma factors regulate expression of the Bacillus subtilis yabE gene via a cis-acting antisense RNA.
J Bacteriol: 2009, 191(3);1101-5
[PubMed:19047346]
[WorldCat.org]
[DOI]
(I p)
Warawan Eiamphungporn, John D Helmann
The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses.
Mol Microbiol: 2008, 67(4);830-48
[PubMed:18179421]
[WorldCat.org]
[DOI]
(P p)
Dirk Höper, Uwe Völker, Michael Hecker
Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis.
J Bacteriol: 2005, 187(8);2810-26
[PubMed:15805528]
[WorldCat.org]
[DOI]
(P p)
Min Cao, Phil A Kobel, Maud M Morshedi, Ming Fang Winston Wu, Chris Paddon, John D Helmann
Defining the Bacillus subtilis sigma(W) regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches.
J Mol Biol: 2002, 316(3);443-57
[PubMed:11866510]
[WorldCat.org]
[DOI]
(P p)