YbgE
- Description: branched-chain amino acid aminotransferase
| Gene name | ybgE |
| Synonyms | |
| Essential | no |
| Product | branched-chain amino acid aminotransferase |
| Function | biosynthesis of branched-chain amino acids |
| Metabolic function and regulation of this protein in SubtiPathways: Lipid synthesis, Ile, Leu, Val, Coenzyme A | |
| MW, pI | 39 kDa, 4.952 |
| Gene length, protein length | 1068 bp, 356 aa |
| Immediate neighbours | ybgB, ybgF |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 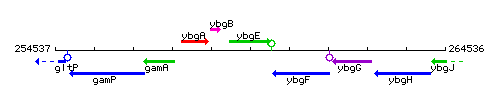
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU02390
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: l-leucine + 2-oxoglutarate = 4-methyl-2-oxopentanoate + L-glutamate (according to Swiss-Prot)
- Protein family: class-IV pyridoxal-phosphate-dependent aminotransferase family (according to Swiss-Prot)
- Paralogous protein(s): YwaA
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: O31461
- KEGG entry: [3]
- E.C. number: 2.6.1.42
Additional information
Expression and regulation
- Operon: ybgE PubMed
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Boris R Belitsky, Abraham L Sonenshein
Roadblock repression of transcription by Bacillus subtilis CodY.
J Mol Biol: 2011, 411(4);729-43
[PubMed:21699902]
[WorldCat.org]
[DOI]
(I p)
Boris R Belitsky, Abraham L Sonenshein
Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis.
J Bacteriol: 2008, 190(4);1224-36
[PubMed:18083814]
[WorldCat.org]
[DOI]
(I p)
Ulrike Mäder, Susanne Hennig, Michael Hecker, Georg Homuth
Transcriptional organization and posttranscriptional regulation of the Bacillus subtilis branched-chain amino acid biosynthesis genes.
J Bacteriol: 2004, 186(8);2240-52
[PubMed:15060025]
[WorldCat.org]
[DOI]
(P p)
Bradley J Berger, Shane English, Gene Chan, Marvin H Knodel
Methionine regeneration and aminotransferases in Bacillus subtilis, Bacillus cereus, and Bacillus anthracis.
J Bacteriol: 2003, 185(8);2418-31
[PubMed:12670965]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Yoshiko Nakaura, Robert P Shivers, Hirotake Yamaguchi, Richard Losick, Yasutaro Fujita, Abraham L Sonenshein
Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis.
J Bacteriol: 2003, 185(6);1911-22
[PubMed:12618455]
[WorldCat.org]
[DOI]
(P p)
Ulrike Mäder, Georg Homuth, Christian Scharf, Knut Büttner, Rüdiger Bode, Michael Hecker
Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability.
J Bacteriol: 2002, 184(15);4288-95
[PubMed:12107147]
[WorldCat.org]
[DOI]
(P p)