Difference between revisions of "Xpt"
| Line 24: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[pbuX]]'', ''[[ypwA]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[pbuX]]'', ''[[ypwA]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU22070 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU22070 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU22070 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU22070 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU22070 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU22070 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:xpt_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:xpt_context.gif]] | ||
Revision as of 10:32, 14 May 2013
- Description: xanthine phosphoribosyltransferase
| Gene name | xpt |
| Synonyms | |
| Essential | no |
| Product | xanthine phosphoribosyltransferase |
| Function | purine salvage and interconversion |
| Gene expression levels in SubtiExpress: xpt | |
| Metabolic function and regulation of this protein in SubtiPathways: Purine salvage, Nucleotides (regulation) | |
| MW, pI | 20 kDa, 5.835 |
| Gene length, protein length | 582 bp, 194 aa |
| Immediate neighbours | pbuX, ypwA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 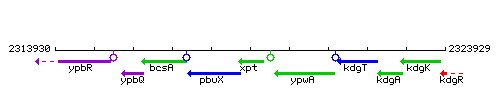
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed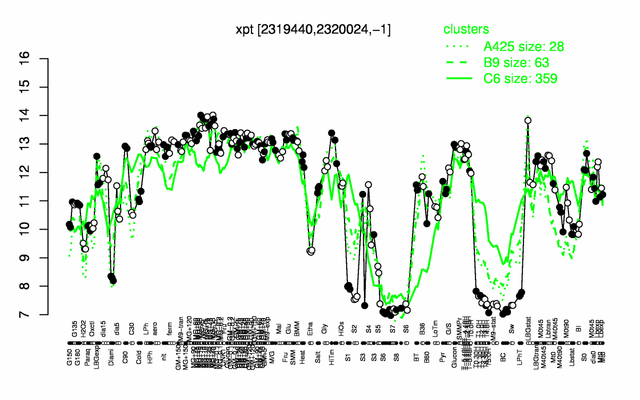
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of nucleotides, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22070
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: XMP + diphosphate = 5-phospho-alpha-D-ribose 1-diphosphate + xanthine (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
Database entries
- UniProt: P42085
- KEGG entry: [3]
- E.C. number: 2.4.2.22
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulation:
- Regulatory mechanism:
- G-box: transcription termination/ antitermination (riboswitch) PubMed
- PurR: transcription repression (molecular inducer: PRPP) PubMed
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant:
- Expression vector:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Otmar M Ottink, Ivo M Westerweele, Marco Tesssari, Frank H T Nelissen, Hans A Heus, Sybren S Wijmenga
1H and 13C resonance assignments of a guanine sensing riboswitch's terminator hairpin.
Biomol NMR Assign: 2010, 4(1);89-91
[PubMed:20204714]
[WorldCat.org]
[DOI]
(I p)
Janina Buck, Jonas Noeske, Jens Wöhnert, Harald Schwalbe
Dissecting the influence of Mg2+ on 3D architecture and ligand-binding of the guanine-sensing riboswitch aptamer domain.
Nucleic Acids Res: 2010, 38(12);4143-53
[PubMed:20200045]
[WorldCat.org]
[DOI]
(I p)
Susan Arent, Anders Kadziola, Sine Larsen, Jan Neuhard, Kaj Frank Jensen
The extraordinary specificity of xanthine phosphoribosyltransferase from Bacillus subtilis elucidated by reaction kinetics, ligand binding, and crystallography.
Biochemistry: 2006, 45(21);6615-27
[PubMed:16716072]
[WorldCat.org]
[DOI]
(P p)
Per Nygaard, Hans H Saxild
The purine efflux pump PbuE in Bacillus subtilis modulates expression of the PurR and G-box (XptR) regulons by adjusting the purine base pool size.
J Bacteriol: 2005, 187(2);791-4
[PubMed:15629952]
[WorldCat.org]
[DOI]
(P p)
Alexander Serganov, Yu-Ren Yuan, Olga Pikovskaya, Anna Polonskaia, Lucy Malinina, Anh Tuân Phan, Claudia Hobartner, Ronald Micura, Ronald R Breaker, Dinshaw J Patel
Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs.
Chem Biol: 2004, 11(12);1729-41
[PubMed:15610857]
[WorldCat.org]
[DOI]
(P p)
Lars Engholm Johansen, Per Nygaard, Catharina Lassen, Yvonne Agersø, Hans H Saxild
Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL).
J Bacteriol: 2003, 185(17);5200-9
[PubMed:12923093]
[WorldCat.org]
[DOI]
(P p)
H H Saxild, K Brunstedt, K I Nielsen, H Jarmer, P Nygaard
Definition of the Bacillus subtilis PurR operator using genetic and bioinformatic tools and expansion of the PurR regulon with glyA, guaC, pbuG, xpt-pbuX, yqhZ-folD, and pbuO.
J Bacteriol: 2001, 183(21);6175-83
[PubMed:11591660]
[WorldCat.org]
[DOI]
(P p)
L C Christiansen, S Schou, P Nygaard, H H Saxild
Xanthine metabolism in Bacillus subtilis: characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism.
J Bacteriol: 1997, 179(8);2540-50
[PubMed:9098051]
[WorldCat.org]
[DOI]
(P p)