Difference between revisions of "Xpt"
(→Expression and regulation) |
|||
| Line 91: | Line 91: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''[[xpt]]-[[pbuX]]'' | + | * '''Operon:''' ''[[xpt]]-[[pbuX]]'' {{PubMed|9098051}} |
| − | * '''[[Sigma factor]]:''' | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|9098051}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| − | |||
** repressed in the presence of adenine or adenosine ([[PurR]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/11591660 PubMed] | ** repressed in the presence of adenine or adenosine ([[PurR]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/11591660 PubMed] | ||
** induced in the absence of guanine ([[G-box]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/12787499 PubMed] | ** induced in the absence of guanine ([[G-box]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/12787499 PubMed] | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | + | ** [[G-box]]: transcription termination/ antitermination ([[riboswitch]]) {{PubMed|9098051,12787499}} | |
| − | ** [[G-box]]: transcription termination/ antitermination ([[riboswitch]]) | ||
** [[PurR]]: transcription repression (molecular inducer: PRPP) [http://www.ncbi.nlm.nih.gov/sites/entrez/11591660 PubMed] | ** [[PurR]]: transcription repression (molecular inducer: PRPP) [http://www.ncbi.nlm.nih.gov/sites/entrez/11591660 PubMed] | ||
| − | * '''Additional information:''' subject to Clp-dependent proteolysis upon glucose starvation | + | * '''Additional information:''' subject to Clp-dependent proteolysis upon glucose starvation {{PubMed|17981983}} |
=Biological materials = | =Biological materials = | ||
Revision as of 20:23, 26 October 2009
- Description: xanthine phosphoribosyltransferase
| Gene name | xpt |
| Synonyms | |
| Essential | no |
| Product | xanthine phosphoribosyltransferase |
| Function | purine salvage and interconversion |
| Metabolic function and regulation of this protein in SubtiPathways: Purine salvage, Nucleotides (regulation) | |
| MW, pI | 20 kDa, 5.835 |
| Gene length, protein length | 582 bp, 194 aa |
| Immediate neighbours | pbuX, ypwA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 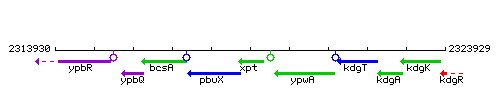
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU22070
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: XMP + diphosphate = 5-phospho-alpha-D-ribose 1-diphosphate + xanthine (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- UniProt: P42085
- KEGG entry: [3]
- E.C. number: 2.4.2.22
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulation:
- Regulatory mechanism:
- G-box: transcription termination/ antitermination (riboswitch) PubMed
- PurR: transcription repression (molecular inducer: PRPP) PubMed
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Susan Arent, Anders Kadziola, Sine Larsen, Jan Neuhard, Kaj Frank Jensen
The extraordinary specificity of xanthine phosphoribosyltransferase from Bacillus subtilis elucidated by reaction kinetics, ligand binding, and crystallography.
Biochemistry: 2006, 45(21);6615-27
[PubMed:16716072]
[WorldCat.org]
[DOI]
(P p)
Per Nygaard, Hans H Saxild
The purine efflux pump PbuE in Bacillus subtilis modulates expression of the PurR and G-box (XptR) regulons by adjusting the purine base pool size.
J Bacteriol: 2005, 187(2);791-4
[PubMed:15629952]
[WorldCat.org]
[DOI]
(P p)
Alexander Serganov, Yu-Ren Yuan, Olga Pikovskaya, Anna Polonskaia, Lucy Malinina, Anh Tuân Phan, Claudia Hobartner, Ronald Micura, Ronald R Breaker, Dinshaw J Patel
Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs.
Chem Biol: 2004, 11(12);1729-41
[PubMed:15610857]
[WorldCat.org]
[DOI]
(P p)
Lars Engholm Johansen, Per Nygaard, Catharina Lassen, Yvonne Agersø, Hans H Saxild
Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL).
J Bacteriol: 2003, 185(17);5200-9
[PubMed:12923093]
[WorldCat.org]
[DOI]
(P p)
H H Saxild, K Brunstedt, K I Nielsen, H Jarmer, P Nygaard
Definition of the Bacillus subtilis PurR operator using genetic and bioinformatic tools and expansion of the PurR regulon with glyA, guaC, pbuG, xpt-pbuX, yqhZ-folD, and pbuO.
J Bacteriol: 2001, 183(21);6175-83
[PubMed:11591660]
[WorldCat.org]
[DOI]
(P p)
L C Christiansen, S Schou, P Nygaard, H H Saxild
Xanthine metabolism in Bacillus subtilis: characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism.
J Bacteriol: 1997, 179(8);2540-50
[PubMed:9098051]
[WorldCat.org]
[DOI]
(P p)