Difference between revisions of "XlyB"
| Line 11: | Line 11: | ||
|style="background:#ABCDEF;" align="center"| '''Product''' || N-acetylmuramoyl-L-alanine amidase | |style="background:#ABCDEF;" align="center"| '''Product''' || N-acetylmuramoyl-L-alanine amidase | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || PBSX prophage-mediated lysis | + | |style="background:#ABCDEF;" align="center"|'''Function''' || [[PBSX prophage]]-mediated lysis |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU12460 xlyB] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU12460 xlyB] | ||
| Line 34: | Line 34: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
{{SubtiWiki category|[[cell wall degradation/ turnover]]}}, | {{SubtiWiki category|[[cell wall degradation/ turnover]]}}, | ||
| − | {{SubtiWiki category|[[phage-related functions]]}} | + | {{SubtiWiki category|[[phage-related functions]]}}, |
| + | {{SubtiWiki category|[[PBSX prophage]]}} | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| Line 62: | Line 59: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 111: | Line 105: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=xlyB_1317535_1318488_1 xlyB] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=xlyB_1317535_1318488_1 xlyB] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' | * '''Regulation:''' | ||
Revision as of 20:12, 28 April 2013
- Description: N-acetylmuramoyl-L-alanine amidase
| Gene name | xlyB |
| Synonyms | yjpB |
| Essential | no |
| Product | N-acetylmuramoyl-L-alanine amidase |
| Function | PBSX prophage-mediated lysis |
| Gene expression levels in SubtiExpress: xlyB | |
| MW, pI | 33 kDa, 9.802 |
| Gene length, protein length | 951 bp, 317 aa |
| Immediate neighbours | yjpA, yjqA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 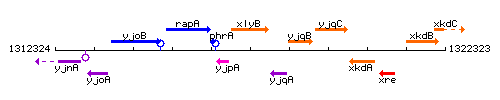
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell wall degradation/ turnover, phage-related functions, PBSX prophage
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU12460
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides (according to Swiss-Prot)
- Protein family: LysM repeat (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- secreted (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: O34391
- KEGG entry: [2]
- E.C. number: 3.5.1.28
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information: the mRNA is very stable (half-life > 15 min) PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References