Difference between revisions of "WapA"
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || unknown | |style="background:#ABCDEF;" align="center"|'''Function''' || unknown | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU39230 wapA] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 258 kDa, 6.131 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 258 kDa, 6.131 | ||
Revision as of 17:29, 7 August 2012
- Description: cell wall-associated protein precursor
| Gene name | wapA |
| Synonyms | |
| Essential | no |
| Product | cell wall-associated protein precursor |
| Function | unknown |
| Gene expression levels in SubtiExpress: wapA | |
| MW, pI | 258 kDa, 6.131 |
| Gene length, protein length | 7002 bp, 2334 aa |
| Immediate neighbours | yxxG, yxxF |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 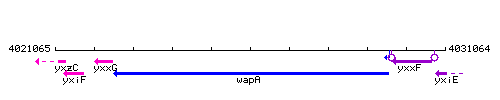
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed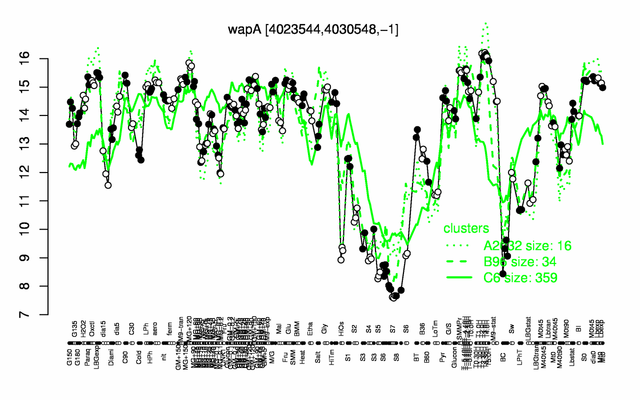
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU39230
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relexed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains: cell wall binding domain as retention signal
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: extracellular (signal peptide), cell wall binding domain as retention signal, major constituent of the secretome PubMed
Database entries
- Structure:
- UniProt: Q07833
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Christopher T Brown, Laura K Fishwick, Binna M Chokshi, Marissa A Cuff, Jay M Jackson, Travis Oglesby, Alison T Rioux, Enrique Rodriguez, Gregory S Stupp, Austin H Trupp, James S Woollcombe-Clarke, Tracy N Wright, William J Zaragoza, Jennifer C Drew, Eric W Triplett, Wayne L Nicholson
Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation.
Appl Environ Microbiol: 2011, 77(19);6867-77
[PubMed:21821766]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Juliane Ollinger, Kyung-Bok Song, Haike Antelmann, Michael Hecker, John D Helmann
Role of the Fur regulon in iron transport in Bacillus subtilis.
J Bacteriol: 2006, 188(10);3664-73
[PubMed:16672620]
[WorldCat.org]
[DOI]
(P p)
Masakuni Serizawa, Keisuke Kodama, Hiroki Yamamoto, Kazuo Kobayashi, Naotake Ogasawara, Junichi Sekiguchi
Functional analysis of the YvrGHb two-component system of Bacillus subtilis: identification of the regulated genes by DNA microarray and northern blot analyses.
Biosci Biotechnol Biochem: 2005, 69(11);2155-69
[PubMed:16306698]
[WorldCat.org]
[DOI]
(P p)
Haike Antelmann, Hiroki Yamamoto, Junichi Sekiguchi, Michael Hecker
Stabilization of cell wall proteins in Bacillus subtilis: a proteomic approach.
Proteomics: 2002, 2(5);591-602
[PubMed:11987133]
[WorldCat.org]
[DOI]
(P p)
V Dartois, M Débarbouillé, F Kunst, G Rapoport
Characterization of a novel member of the DegS-DegU regulon affected by salt stress in Bacillus subtilis.
J Bacteriol: 1998, 180(7);1855-61
[PubMed:9537385]
[WorldCat.org]
[DOI]
(P p)