Difference between revisions of "UvrC"
(→References) |
(→The protein) |
||
| Line 72: | Line 72: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''Interactions:''' [[UvrB]]-[[UvrC]] {{PubMed|16464004}} |
* '''Localization:''' | * '''Localization:''' | ||
| Line 78: | Line 78: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=3C65 3C65] ( | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=3C65 3C65] (5' endonuclease domain, Geobacillus stearothermophilus) |
* '''UniProt:''' [http://www.uniprot.org/uniprot/P14951 P14951] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P14951 P14951] | ||
Revision as of 12:37, 14 December 2009
- Description: excinuclease ABC (subunit C)
| Gene name | uvrC |
| Synonyms | uvrB |
| Essential | no |
| Product | excinuclease ABC (subunit C) |
| Function | DNA repair after UV damage |
| MW, pI | 69 kDa, 9.503 |
| Gene length, protein length | 1794 bp, 598 aa |
| Immediate neighbours | ask, trxA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 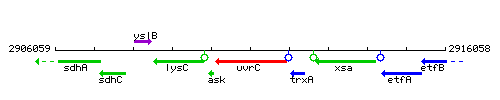
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU28490
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: uvrC family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure: 3C65 (5' endonuclease domain, Geobacillus stearothermophilus)
- UniProt: P14951
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
James J Truglio, Deborah L Croteau, Bennett Van Houten, Caroline Kisker
Prokaryotic nucleotide excision repair: the UvrABC system.
Chem Rev: 2006, 106(2);233-52
[PubMed:16464004]
[WorldCat.org]
[DOI]
(P p)
Bennett Van Houten, Deborah L Croteau, Matthew J DellaVecchia, Hong Wang, Caroline Kisker
'Close-fitting sleeves': DNA damage recognition by the UvrABC nuclease system.
Mutat Res: 2005, 577(1-2);92-117
[PubMed:15927210]
[WorldCat.org]
[DOI]
(P p)
A Sancar
Mechanisms of DNA excision repair.
Science: 1994, 266(5193);1954-6
[PubMed:7801120]
[WorldCat.org]
[DOI]
(P p)
Original Publications
Nora Au, Elke Kuester-Schoeck, Veena Mandava, Laura E Bothwell, Susan P Canny, Karen Chachu, Sierra A Colavito, Shakierah N Fuller, Eli S Groban, Laura A Hensley, Theresa C O'Brien, Amish Shah, Jessica T Tierney, Louise L Tomm, Thomas M O'Gara, Alexi I Goranov, Alan D Grossman, Charles M Lovett
Genetic composition of the Bacillus subtilis SOS system.
J Bacteriol: 2005, 187(22);7655-66
[PubMed:16267290]
[WorldCat.org]
[DOI]
(P p)
J J Lin, A Sancar
Reconstitution of nucleotide excision nuclease with UvrA and UvrB proteins from Escherichia coli and UvrC protein from Bacillus subtilis.
J Biol Chem: 1990, 265(34);21337-41
[PubMed:2174442]
[WorldCat.org]
(P p)
N Y Chen, J J Zhang, H Paulus
Chromosomal location of the Bacillus subtilis aspartokinase II gene and nucleotide sequence of the adjacent genes homologous to uvrC and trx of Escherichia coli.
J Gen Microbiol: 1989, 135(11);2931-40
[PubMed:2559145]
[WorldCat.org]
[DOI]
(P p)