Difference between revisions of "Ung"
| Line 24: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ywdH]]'', ''[[ywdF]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ywdH]]'', ''[[ywdF]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU37970 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU37970 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU37970 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU37970 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU37970 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU37970 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:ung_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:ung_context.gif]] | ||
Revision as of 11:35, 14 May 2013
- Description: uracil-DNA glycosylase
| Gene name | ung |
| Synonyms | ipa-57d, ywdG |
| Essential | no |
| Product | uracil-DNA glycosylase |
| Function | DNA repair |
| Gene expression levels in SubtiExpress: ung | |
| Interactions involving this protein in SubtInteract: Ung | |
| MW, pI | 25 kDa, 8.782 |
| Gene length, protein length | 675 bp, 225 aa |
| Immediate neighbours | ywdH, ywdF |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 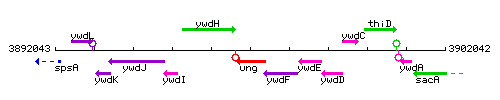
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU37970
Phenotypes of a mutant
- increased mutation rates PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: removes uracil preferentially from single-stranded DNA over double-stranded DNA, exhibiting higher preference for U:G than U:A mismatches PubMed
- Protein family: uracil-DNA glycosylase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- UniProt: P39615
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: ung PubMed
- Sigma factor:
- Regulation:
- expressed throughout growth and staionary phase PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Karina López-Olmos, Martha P Hernández, Jorge A Contreras-Garduño, Eduardo A Robleto, Peter Setlow, Ronald E Yasbin, Mario Pedraza-Reyes
Roles of endonuclease V, uracil-DNA glycosylase, and mismatch repair in Bacillus subtilis DNA base-deamination-induced mutagenesis.
J Bacteriol: 2012, 194(2);243-52
[PubMed:22056936]
[WorldCat.org]
[DOI]
(I p)
Prem Singh Kaushal, Ramappa K Talawar, Umesh Varshney, M Vijayan
Structure of uracil-DNA glycosylase from Mycobacterium tuberculosis: insights into interactions with ligands.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2010, 66(Pt 8);887-92
[PubMed:20693660]
[WorldCat.org]
[DOI]
(I p)
E Presecan, I Moszer, L Boursier, H Cruz Ramos, V de la Fuente, M-F Hullo, C Lelong, S Schleich, A Sekowska, B H Song, G Villani, F Kunst, A Danchin, P Glaser
The Bacillus subtilis genome from gerBC (311 degrees) to licR (334 degrees).
Microbiology (Reading): 1997, 143 ( Pt 10);3313-3328
[PubMed:9353933]
[WorldCat.org]
[DOI]
(P p)