Difference between revisions of "TyrZ"
| Line 58: | Line 58: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU38460&redirect=T BSU38460] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/tyrZ.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/tyrZ.html] | ||
| Line 96: | Line 97: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU38460&redirect=T BSU38460] | ||
* '''Structure:''' | * '''Structure:''' | ||
Revision as of 16:08, 2 April 2014
- Description: tyrosyl-tRNA synthetase (minor)
| Gene name | tyrZ |
| Synonyms | ipa-9r, tyrS1, tyrT |
| Essential | no |
| Product | tyrosyl-tRNA synthetase (minor) |
| Function | translation |
| Gene expression levels in SubtiExpress: tyrZ | |
| Metabolic function and regulation of this protein in SubtiPathways: tyrZ | |
| MW, pI | 46 kDa, 6.661 |
| Gene length, protein length | 1239 bp, 413 aa |
| Immediate neighbours | ywaE, ywaD |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 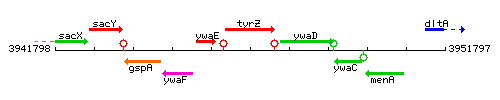
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed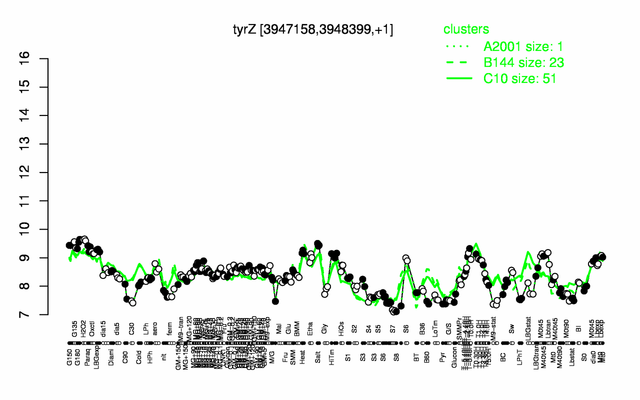
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU38460
Phenotypes of a mutant
Database entries
- BsubCyc: BSU38460
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + L-tyrosine + tRNA(Tyr) = AMP + diphosphate + L-tyrosyl-tRNA(Tyr) (according to Swiss-Prot) ATP + L-tyrosine + tRNA(Tyr) = AMP + diphosphate + L-tyrosyl-tRNA(Tyr) (according to Swiss-Prot)
- Protein family: TyrS type 2 subfamily (according to Swiss-Prot), class-I aminoacyl-tRNA synthetase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylated on Thr-311 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU38460
- Structure:
- UniProt: P25151
- KEGG entry: [3]
- E.C. number: 6.1.1.1
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- T-box: RNA switch, transcriptional antitermination PubMed
- Additional information:
Biological materials
- Mutant: QB53532 (aphA3), available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Boumediene Soufi, Chanchal Kumar, Florian Gnad, Matthias Mann, Ivan Mijakovic, Boris Macek
Stable isotope labeling by amino acids in cell culture (SILAC) applied to quantitative proteomics of Bacillus subtilis.
J Proteome Res: 2010, 9(7);3638-46
[PubMed:20509597]
[WorldCat.org]
[DOI]
(I p)
Ana Gutiérrez-Preciado, Tina M Henkin, Frank J Grundy, Charles Yanofsky, Enrique Merino
Biochemical features and functional implications of the RNA-based T-box regulatory mechanism.
Microbiol Mol Biol Rev: 2009, 73(1);36-61
[PubMed:19258532]
[WorldCat.org]
[DOI]
(I p)
T M Henkin, B L Glass, F J Grundy
Analysis of the Bacillus subtilis tyrS gene: conservation of a regulatory sequence in multiple tRNA synthetase genes.
J Bacteriol: 1992, 174(4);1299-306
[PubMed:1735721]
[WorldCat.org]
[DOI]
(P p)