Difference between revisions of "TyrA"
| Line 54: | Line 54: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| Line 80: | Line 78: | ||
* '''Effectors of protein activity:''' subject to feedback inhibtion by tyrosine {{PubMed|4956345}} | * '''Effectors of protein activity:''' subject to feedback inhibtion by tyrosine {{PubMed|4956345}} | ||
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| − | * '''Localization:''' cytoplasm (according to Swiss-Prot) | + | * '''[[Localization]]:''' |
| + | ** cytoplasm (according to Swiss-Prot) | ||
=== Database entries === | === Database entries === | ||
| Line 106: | Line 105: | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| − | ** induction by tryptophan ([[MtrB| TRAP]]) {{PubMed|1551827}} | + | ** induction by tryptophan ([[MtrB|TRAP]]) {{PubMed|1551827}} |
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | ** [[MtrB| TRAP]]: binding to the mRNA in the presence of tryptophan, this results in transcription termination {{PubMed|8419914}} | + | ** [[MtrB|TRAP]]: binding to the mRNA in the presence of tryptophan, this results in transcription termination {{PubMed|8419914}} |
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** the mRNA is substantially stabilized upon depletion of [[Rny|RNase Y]] {{PubMed|21815947}} | ||
=Biological materials = | =Biological materials = | ||
| Line 132: | Line 132: | ||
=References= | =References= | ||
| − | + | <big>''Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J'' </big> | |
| + | <big>'''RNA processing in ''Bacillus subtilis'': identification of targets of the essential RNase Y.''' </big> | ||
| + | <big>Mol Microbiol. 2011 81(6): 1459-1473. </big> | ||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/21815947 PubMed:21815947] | ||
<pubmed>,3106153, 4956345 3924737 6436812 1551827 8419914</pubmed> | <pubmed>,3106153, 4956345 3924737 6436812 1551827 8419914</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 18:29, 19 November 2011
- Description: prephenate dehydrogenase
| Gene name | tyrA |
| Synonyms | |
| Essential | no |
| Product | prephenate dehydrogenase |
| Function | biosynthesis of tyrosine |
| Metabolic function and regulation of this protein in SubtiPathways: Phe, Tyr, Trp | |
| MW, pI | 41 kDa, 5.471 |
| Gene length, protein length | 1113 bp, 371 aa |
| Immediate neighbours | aroE, hisC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 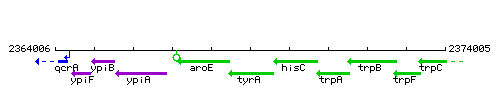
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22610
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Prephenate + NAD+ = 4-hydroxyphenylpyruvate + CO2 + NADH (according to Swiss-Prot)
- Protein family: HisMQ subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: subject to feedback inhibtion by tyrosine PubMed
- Localization:
- cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P20692
- KEGG entry: [3]
- E.C. number: 1.3.1.12
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol Microbiol. 2011 81(6): 1459-1473. PubMed:21815947
J Otridge, P Gollnick
MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan-dependent manner.
Proc Natl Acad Sci U S A: 1993, 90(1);128-32
[PubMed:8419914]
[WorldCat.org]
[DOI]
(P p)
P Babitzke, P Gollnick, C Yanofsky
The mtrAB operon of Bacillus subtilis encodes GTP cyclohydrolase I (MtrA), an enzyme involved in folic acid biosynthesis, and MtrB, a regulator of tryptophan biosynthesis.
J Bacteriol: 1992, 174(7);2059-64
[PubMed:1551827]
[WorldCat.org]
[DOI]
(P p)
D J Henner, L Band, G Flaggs, E Chen
The organization and nucleotide sequence of the Bacillus subtilis hisH, tyrA and aroE genes.
Gene: 1986, 49(1);147-52
[PubMed:3106153]
[WorldCat.org]
[DOI]
(P p)
D J Henner, L Band, H Shimotsu
Nucleotide sequence of the Bacillus subtilis tryptophan operon.
Gene: 1985, 34(2-3);169-77
[PubMed:3924737]
[WorldCat.org]
[DOI]
(P p)
H Shimotsu, D J Henner
Characterization of the Bacillus subtilis tryptophan promoter region.
Proc Natl Acad Sci U S A: 1984, 81(20);6315-9
[PubMed:6436812]
[WorldCat.org]
[DOI]
(P p)
E W Nester, R A Jensen
Control of aromatic acid biosynthesis in Bacillus subtilis: sequenial feedback inhibition.
J Bacteriol: 1966, 91(4);1594-8
[PubMed:4956345]
[WorldCat.org]
[DOI]
(P p)