Difference between revisions of "TufA"
| Line 37: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 87: | Line 83: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
| − | * '''Modification:''' phosphorylated on Thr-384 by [[PrkC]], dephosphorylated by [[PrpC]] [http://www.ncbi.nlm.nih.gov/sites/entrez/19246764 PubMed], [http://www.ncbi.nlm.nih.gov/pubmed/17726680 PubMed] | + | * '''Modification:''' |
| + | ** phosphorylated on Thr-384 by [[PrkC]], dephosphorylated by [[PrpC]] [http://www.ncbi.nlm.nih.gov/sites/entrez/19246764 PubMed], [http://www.ncbi.nlm.nih.gov/pubmed/17726680 PubMed] | ||
** Cys83 is S-bacillithiolated in B. subtilis and other Bacillus species [http://www.ncbi.nlm.nih.gov/pubmed/22938038 PubMed] | ** Cys83 is S-bacillithiolated in B. subtilis and other Bacillus species [http://www.ncbi.nlm.nih.gov/pubmed/22938038 PubMed] | ||
| + | ** phosphorylated on several Arg residues {{PubMed|24263382}} | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 121: | Line 119: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=tufA_132882_134072_1 tufA] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=tufA_132882_134072_1 tufA] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 154: | Line 152: | ||
=References= | =References= | ||
| − | <pubmed>22938038,19246764,20133608 , 19246764, 17726680, 18763711 </pubmed> | + | <pubmed>22938038,19246764,20133608 , 19246764, 17726680, 18763711 24263382 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:52, 17 December 2013
- Description: elongation factor Tu
| Gene name | tufA |
| Synonyms | |
| Essential | yes PubMed |
| Product | elongation factor Tu |
| Function | translation |
| Gene expression levels in SubtiExpress: tufA | |
| Interactions involving this protein in SubtInteract: TufA | |
| MW, pI | 43 kDa, 4.723 |
| Gene length, protein length | 1188 bp, 396 aa |
| Immediate neighbours | fusA, ybaC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 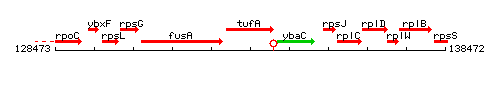
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
translation, essential genes, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01130
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: EF-Tu/EF-1A subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P33166
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Andreas Schmidt, Débora Broch Trentini, Silvia Spiess, Jakob Fuhrmann, Gustav Ammerer, Karl Mechtler, Tim Clausen
Quantitative phosphoproteomics reveals the role of protein arginine phosphorylation in the bacterial stress response.
Mol Cell Proteomics: 2014, 13(2);537-50
[PubMed:24263382]
[WorldCat.org]
[DOI]
(I p)
Bui Khanh Chi, Alexandra A Roberts, Tran Thi Thanh Huyen, Katrin Bäsell, Dörte Becher, Dirk Albrecht, Chris J Hamilton, Haike Antelmann
S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria.
Antioxid Redox Signal: 2013, 18(11);1273-95
[PubMed:22938038]
[WorldCat.org]
[DOI]
(I p)
Hervé Joël Defeu Soufo, Christian Reimold, Uwe Linne, Tobias Knust, Johannes Gescher, Peter L Graumann
Bacterial translation elongation factor EF-Tu interacts and colocalizes with actin-like MreB protein.
Proc Natl Acad Sci U S A: 2010, 107(7);3163-8
[PubMed:20133608]
[WorldCat.org]
[DOI]
(I p)
Cédric Absalon, Michal Obuchowski, Edwige Madec, Delphine Delattre, I Barry Holland, Simone J Séror
CpgA, EF-Tu and the stressosome protein YezB are substrates of the Ser/Thr kinase/phosphatase couple, PrkC/PrpC, in Bacillus subtilis.
Microbiology (Reading): 2009, 155(Pt 3);932-943
[PubMed:19246764]
[WorldCat.org]
[DOI]
(P p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Dörte Becher, Jörg Bernhardt, Katrin Gronau, Anja Klutzny, Michael Hecker
Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis.
Proteomics: 2007, 7(19);3509-26
[PubMed:17726680]
[WorldCat.org]
[DOI]
(P p)