Difference between revisions of "TsaC"
| Line 1: | Line 1: | ||
| − | * '''Description:''' biosynthesis of the hypermodified base threonylcarbamoyladenosine (t(6)A) (together with [[ | + | * '''Description:''' L-threonylcarbamoyl-AMP synthase, biosynthesis of the hypermodified base threonylcarbamoyladenosine (t(6)A) (together with [[TsaB]]-[[TsaD]]-[[TsaE]]) <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || L-threonylcarbamoyl-AMP synthase |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || tRNA modification | |style="background:#ABCDEF;" align="center"|'''Function''' || tRNA modification | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU36950 | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU36950 tsaC] |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 36 kDa, 5.82 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 36 kDa, 5.82 | ||
| Line 35: | Line 35: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 62: | Line 58: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| Line 70: | Line 64: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' biosynthesis of the hypermodified base threonylcarbamoyladenosine (t(6)A), a universal modification found at position 37 of ANN decoding tRNAs, which imparts a unique structure to the anticodon loop enhancing its binding to ribosomes in vitro (shown for the corresponding ''E. coli'' protein YrdC) {{PubMed|19287007}} | + | * '''Catalyzed reaction/ biological activity:''' |
| + | ** conversion of L-threonine, bicarbonate/CO2 and ATP to give the intermediate L-threonylcarbamoyl-AMP (TC-AMP) and pyrophosphate as products {{PubMed|23072323}} | ||
| + | ** biosynthesis of the hypermodified base threonylcarbamoyladenosine (t(6)A), a universal modification found at position 37 of ANN decoding tRNAs, which imparts a unique structure to the anticodon loop enhancing its binding to ribosomes in vitro (shown for the corresponding ''E. coli'' protein YrdC) {{PubMed|19287007}} | ||
* '''Protein family:''' SUA5 family (according to Swiss-Prot) | * '''Protein family:''' SUA5 family (according to Swiss-Prot) | ||
| Line 139: | Line 135: | ||
=References= | =References= | ||
| − | <pubmed>17005971 17932079 19287007 </pubmed> | + | <pubmed>17005971 17932079 19287007 23072323 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:07, 18 October 2012
- Description: L-threonylcarbamoyl-AMP synthase, biosynthesis of the hypermodified base threonylcarbamoyladenosine (t(6)A) (together with TsaB-TsaD-TsaE)
| Gene name | ywlC |
| Synonyms | ipc-29d |
| Essential | no |
| Product | L-threonylcarbamoyl-AMP synthase |
| Function | tRNA modification |
| Gene expression levels in SubtiExpress: tsaC | |
| MW, pI | 36 kDa, 5.82 |
| Gene length, protein length | 1038 bp, 346 aa |
| Immediate neighbours | ywlD, ywlB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 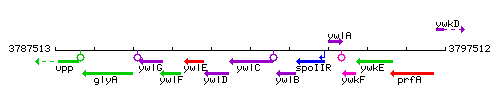
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU36950
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- conversion of L-threonine, bicarbonate/CO2 and ATP to give the intermediate L-threonylcarbamoyl-AMP (TC-AMP) and pyrophosphate as products PubMed
- biosynthesis of the hypermodified base threonylcarbamoyladenosine (t(6)A), a universal modification found at position 37 of ANN decoding tRNAs, which imparts a unique structure to the anticodon loop enhancing its binding to ribosomes in vitro (shown for the corresponding E. coli protein YrdC) PubMed
- Protein family: SUA5 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P39153
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Charles T Lauhon
Mechanism of N6-threonylcarbamoyladenonsine (t(6)A) biosynthesis: isolation and characterization of the intermediate threonylcarbamoyl-AMP.
Biochemistry: 2012, 51(44);8950-63
[PubMed:23072323]
[WorldCat.org]
[DOI]
(I p)
Basma El Yacoubi, Benjamin Lyons, Yulien Cruz, Robert Reddy, Brian Nordin, Fabio Agnelli, James R Williamson, Paul Schimmel, Manal A Swairjo, Valérie de Crécy-Lagard
The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA.
Nucleic Acids Res: 2009, 37(9);2894-909
[PubMed:19287007]
[WorldCat.org]
[DOI]
(I p)
Shu Ishikawa, Yoshitoshi Ogura, Mika Yoshimura, Hajime Okumura, Eunha Cho, Yoshikazu Kawai, Ken Kurokawa, Taku Oshima, Naotake Ogasawara
Distribution of stable DnaA-binding sites on the Bacillus subtilis genome detected using a modified ChIP-chip method.
DNA Res: 2007, 14(4);155-68
[PubMed:17932079]
[WorldCat.org]
[DOI]
(P p)
Alison Hunt, Joy P Rawlins, Helena B Thomaides, Jeff Errington
Functional analysis of 11 putative essential genes in Bacillus subtilis.
Microbiology (Reading): 2006, 152(Pt 10);2895-2907
[PubMed:17005971]
[WorldCat.org]
[DOI]
(P p)