Difference between revisions of "TrpB"
(→Original publications) |
|||
| Line 149: | Line 149: | ||
<pubmed> 18486479 11893063 11163353 7900177 19387555 </pubmed> | <pubmed> 18486479 11893063 11163353 7900177 19387555 </pubmed> | ||
==Original publications== | ==Original publications== | ||
| − | + | <pubmed> 14976255, 3924737, 6436812,2422155,8419914, 1551827, 17507374 21815947 </pubmed> | |
| − | |||
| − | |||
| − | |||
| − | <pubmed> 14976255, 3924737, 6436812,2422155,8419914, 1551827, 17507374 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 16:23, 13 July 2013
- Description: tryptophan synthase (beta subunit)
| Gene name | trpB |
| Synonyms | |
| Essential | no |
| Product | tryptophan synthase (beta subunit) |
| Function | biosynthesis of tryptophan |
| Gene expression levels in SubtiExpress: trpB | |
| Interactions involving this protein in SubtInteract: TrpB | |
| Metabolic function and regulation of this protein in SubtiPathways: Phe, Tyr, Trp | |
| MW, pI | 43 kDa, 5.526 |
| Gene length, protein length | 1200 bp, 400 aa |
| Immediate neighbours | trpA, trpF |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 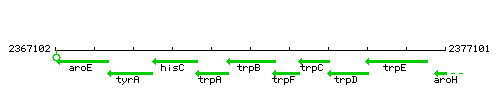
This image was kindly provided by SubtiList
| |
| Expression at a glance PubMed 500px | |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22640
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: L-serine + 1-C-(indol-3-yl)glycerol 3-phosphate = L-tryptophan + glyceraldehyde 3-phosphate + H2O (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P07600
- KEGG entry: [3]
- E.C. number: 4.2.1.20
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Samanta Raboni, Stefano Bettati, Andrea Mozzarelli
Tryptophan synthase: a mine for enzymologists.
Cell Mol Life Sci: 2009, 66(14);2391-403
[PubMed:19387555]
[WorldCat.org]
[DOI]
(I p)
Michael F Dunn, Dimitri Niks, Huu Ngo, Thomas R M Barends, Ilme Schlichting
Tryptophan synthase: the workings of a channeling nanomachine.
Trends Biochem Sci: 2008, 33(6);254-64
[PubMed:18486479]
[WorldCat.org]
[DOI]
(P p)
E W Miles
Tryptophan synthase: a multienzyme complex with an intramolecular tunnel.
Chem Rec: 2001, 1(2);140-51
[PubMed:11893063]
[WorldCat.org]
[DOI]
(P p)
N K Nagradova
Interdomain interactions in oligomeric enzymes: creation of asymmetry in homo-oligomers and role in metabolite channeling between active centers of hetero-oligomers.
FEBS Lett: 2001, 487(3);327-32
[PubMed:11163353]
[WorldCat.org]
[DOI]
(P p)
E W Miles
Tryptophan synthase. Structure, function, and protein engineering.
Subcell Biochem: 1995, 24;207-54
[PubMed:7900177]
[WorldCat.org]
(P p)
Original publications
Martin Lehnik-Habrink, Marc Schaffer, Ulrike Mäder, Christine Diethmaier, Christina Herzberg, Jörg Stülke
RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y.
Mol Microbiol: 2011, 81(6);1459-73
[PubMed:21815947]
[WorldCat.org]
[DOI]
(I p)
Gintaras Deikus, David H Bechhofer
Initiation of decay of Bacillus subtilis trp leader RNA.
J Biol Chem: 2007, 282(28);20238-44
[PubMed:17507374]
[WorldCat.org]
[DOI]
(P p)
Gintaras Deikus, Paul Babitzke, David H Bechhofer
Recycling of a regulatory protein by degradation of the RNA to which it binds.
Proc Natl Acad Sci U S A: 2004, 101(9);2747-51
[PubMed:14976255]
[WorldCat.org]
[DOI]
(P p)
J Otridge, P Gollnick
MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan-dependent manner.
Proc Natl Acad Sci U S A: 1993, 90(1);128-32
[PubMed:8419914]
[WorldCat.org]
[DOI]
(P p)
P Babitzke, P Gollnick, C Yanofsky
The mtrAB operon of Bacillus subtilis encodes GTP cyclohydrolase I (MtrA), an enzyme involved in folic acid biosynthesis, and MtrB, a regulator of tryptophan biosynthesis.
J Bacteriol: 1992, 174(7);2059-64
[PubMed:1551827]
[WorldCat.org]
[DOI]
(P p)
H Shimotsu, M I Kuroda, C Yanofsky, D J Henner
Novel form of transcription attenuation regulates expression the Bacillus subtilis tryptophan operon.
J Bacteriol: 1986, 166(2);461-71
[PubMed:2422155]
[WorldCat.org]
[DOI]
(P p)
D J Henner, L Band, H Shimotsu
Nucleotide sequence of the Bacillus subtilis tryptophan operon.
Gene: 1985, 34(2-3);169-77
[PubMed:3924737]
[WorldCat.org]
[DOI]
(P p)
H Shimotsu, D J Henner
Characterization of the Bacillus subtilis tryptophan promoter region.
Proc Natl Acad Sci U S A: 1984, 81(20);6315-9
[PubMed:6436812]
[WorldCat.org]
[DOI]
(P p)