Difference between revisions of "TrmFO"
| Line 118: | Line 118: | ||
** number of protein molecules per cell (minimal medium with glucose and ammonium): 458 {{PubMed|24696501}} | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 458 {{PubMed|24696501}} | ||
** number of protein molecules per cell (complex medium with amino acids, without glucose): 1069 {{PubMed|24696501}} | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 1069 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 237 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 237 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 439 {{PubMed|21395229}} | ||
=Biological materials = | =Biological materials = | ||
| − | |||
* '''Mutant:''' | * '''Mutant:''' | ||
Revision as of 15:06, 17 April 2014
- Description: flavoprotein, tRNA:m(5)U-54 methyltransferase, glucose-inhibited division protein
| Gene name | trmFO |
| Synonyms | ylyC, gid |
| Essential | no |
| Product | tRNA:m(5)U-54 methyltransferase |
| Function | tRNA modification |
| Gene expression levels in SubtiExpress: trmFO | |
| MW, pI | 47 kDa, 5.767 |
| Gene length, protein length | 1305 bp, 435 aa |
| Immediate neighbours | topA, codV |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 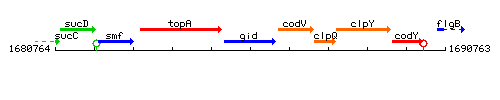
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed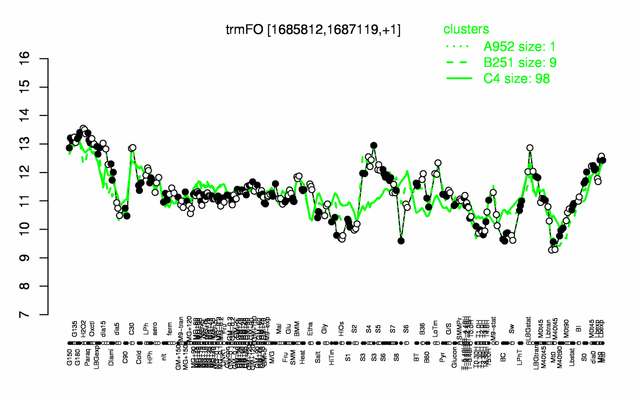
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU16130
Phenotypes of a mutant
Database entries
- BsubCyc: BSU16130
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: catalyzes the folate-dependent C(5)-methylation of uridine at position 54 in the TpsiC loop of tRNA
- Protein family: TrmFO subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU16130
- UniProt: P39815
- KEGG entry: [2]
- E.C. number: 2.1.1.74
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium): 458 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 1069 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 237 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 237 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 439 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Djemel Hamdane, Eduardo Bruch, Sun Un, Martin Field, Marc Fontecave
Activation of a unique flavin-dependent tRNA-methylating agent.
Biochemistry: 2013, 52(49);8949-56
[PubMed:24228791]
[WorldCat.org]
[DOI]
(I p)
Djemel Hamdane, Manuela Argentini, David Cornu, Béatrice Golinelli-Pimpaneau, Marc Fontecave
FAD/folate-dependent tRNA methyltransferase: flavin as a new methyl-transfer agent.
J Am Chem Soc: 2012, 134(48);19739-45
[PubMed:23157377]
[WorldCat.org]
[DOI]
(I p)
Ryota Yamagami, Koki Yamashita, Hiroshi Nishimasu, Chie Tomikawa, Anna Ochi, Chikako Iwashita, Akira Hirata, Ryuichiro Ishitani, Osamu Nureki, Hiroyuki Hori
The tRNA recognition mechanism of folate/FAD-dependent tRNA methyltransferase (TrmFO).
J Biol Chem: 2012, 287(51);42480-94
[PubMed:23095745]
[WorldCat.org]
[DOI]
(I p)
Djemel Hamdane, Manuela Argentini, David Cornu, Hannu Myllykallio, Stéphane Skouloubris, Gaston Hui-Bon-Hoa, Béatrice Golinelli-Pimpaneau
Insights into folate/FAD-dependent tRNA methyltransferase mechanism: role of two highly conserved cysteines in catalysis.
J Biol Chem: 2011, 286(42);36268-80
[PubMed:21846722]
[WorldCat.org]
[DOI]
(I p)
Djemel Hamdane, Vincent Guerineau, Sun Un, Beatrice Golinelli-Pimpaneau
A catalytic intermediate and several flavin redox states stabilized by folate-dependent tRNA methyltransferase from Bacillus subtilis.
Biochemistry: 2011, 50(23);5208-19
[PubMed:21561081]
[WorldCat.org]
[DOI]
(I p)
Djemel Hamdane, Stéphane Skouloubris, Hannu Myllykallio, Béatrice Golinelli-Pimpaneau
Expression and purification of untagged and histidine-tagged folate-dependent tRNA:m5U54 methyltransferase from Bacillus subtilis.
Protein Expr Purif: 2010, 73(1);83-9
[PubMed:20412857]
[WorldCat.org]
[DOI]
(I p)
Hiroshi Nishimasu, Ryuichiro Ishitani, Koki Yamashita, Chikako Iwashita, Akira Hirata, Hiroyuki Hori, Osamu Nureki
Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase.
Proc Natl Acad Sci U S A: 2009, 106(20);8180-5
[PubMed:19416846]
[WorldCat.org]
[DOI]
(I p)
Jaunius Urbonavicius, Stéphane Skouloubris, Hannu Myllykallio, Henri Grosjean
Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria--evolutionary implications.
Nucleic Acids Res: 2005, 33(13);3955-64
[PubMed:16027442]
[WorldCat.org]
[DOI]
(I e)