Difference between revisions of "TnrA"
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' ||regulation of nitrogen assimilation <br/>(positive regulation of nrgAB, nasBCDEF, gabP,<br/> ureABC, guaD; negative regulation of glnRA, gltAB) | |style="background:#ABCDEF;" align="center"|'''Function''' ||regulation of nitrogen assimilation <br/>(positive regulation of nrgAB, nasBCDEF, gabP,<br/> ureABC, guaD; negative regulation of glnRA, gltAB) | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/fatty_acid_syn.html Lipid synthesis], [http://subtiwiki.uni-goettingen.de/pathways/fatty_acid_syn.html Lipid synthesis], [http://subtiwiki.uni-goettingen.de/pathways/gene_regulation_nucleotides.html Nucleotides (regulation)], [http://subtiwiki.uni-goettingen.de/pathways/ile_val_leu.html Ile, Leu, Val], [http://subtiwiki.uni-goettingen.de/pathways/glutamate.html Ammonium/ glutamate], | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/fatty_acid_syn.html Lipid synthesis], [http://subtiwiki.uni-goettingen.de/pathways/fatty_acid_syn.html Lipid synthesis], [http://subtiwiki.uni-goettingen.de/pathways/gene_regulation_nucleotides.html Nucleotides (regulation)], [http://subtiwiki.uni-goettingen.de/pathways/ile_val_leu.html Ile, Leu, Val], <br/>[http://subtiwiki.uni-goettingen.de/pathways/glutamate.html Ammonium/ glutamate], [http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism], [http://subtiwiki.uni-goettingen.de/pathways/cellwall.html Cell wall], <br/>[http://subtiwiki.uni-goettingen.de/pathways/CoA_synthesis.html Coenzyme A], [http://subtiwiki.uni-goettingen.de/pathways/phosphorelay.html Phosphorelay]''' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 12 kDa, 10.235 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 12 kDa, 10.235 | ||
| Line 31: | Line 31: | ||
__TOC__ | __TOC__ | ||
| − | + | <br/><br/><br/><br/><br/> | |
=The gene= | =The gene= | ||
Revision as of 15:28, 16 February 2010
- Description: transcriptional pleiotropic regulator invoved in global nitrogen regulation
| Gene name | tnrA |
| Synonyms | scgR |
| Essential | no |
| Product | transcription activator/ repressor |
| Function | regulation of nitrogen assimilation (positive regulation of nrgAB, nasBCDEF, gabP, ureABC, guaD; negative regulation of glnRA, gltAB) |
| Metabolic function and regulation of this protein in SubtiPathways: Lipid synthesis, Lipid synthesis, Nucleotides (regulation), Ile, Leu, Val, Ammonium/ glutamate, Central C-metabolism, Cell wall, Coenzyme A, Phosphorelay | |
| MW, pI | 12 kDa, 10.235 |
| Gene length, protein length | 330 bp, 110 aa |
| Immediate neighbours | mgtE, ykzB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 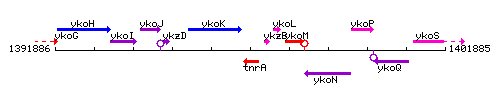
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU13310
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Genes/ operons controlled by TnrA
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: feedback-inhibited GlnA prevents TnrA from DNA binding
- Interactions: TnrA-GlnA, TnrA-NrgB PubMed, TnrA-GlnA, this interaction results in loss of TnrA DNA-binding activity
- Localization:
Database entries
- Structure:
- UniProt: Q45666
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: tnrA (according to DBTBS)
- Regulation:
- Additional information:
Biological materials
- Mutant: GP252 (in frame deletion), available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody: available in the Karl Forchhammer lab
Labs working on this gene/protein
Susan Fisher, Boston, USA homepage
Your additional remarks
References
Reviews
Original publications